| Full name: intraflagellar transport 122 | Alias Symbol: WDR140|WDR10p|SPG | ||
| Type: protein-coding gene | Cytoband: 3q21.3-q22.1 | ||
| Entrez ID: 55764 | HGNC ID: HGNC:13556 | Ensembl Gene: ENSG00000163913 | OMIM ID: 606045 |
| Drug and gene relationship at DGIdb | |||
Expression of IFT122:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | IFT122 | 55764 | 220744_s_at | 0.6950 | 0.1535 | |
| GSE20347 | IFT122 | 55764 | 216678_at | 0.0531 | 0.6692 | |
| GSE23400 | IFT122 | 55764 | 216678_at | -0.1478 | 0.0006 | |
| GSE26886 | IFT122 | 55764 | 220744_s_at | 0.4884 | 0.0649 | |
| GSE29001 | IFT122 | 55764 | 216678_at | -0.2879 | 0.1136 | |
| GSE38129 | IFT122 | 55764 | 220744_s_at | 0.4237 | 0.0048 | |
| GSE45670 | IFT122 | 55764 | 220744_s_at | 0.0698 | 0.7077 | |
| GSE53622 | IFT122 | 55764 | 54091 | 0.4475 | 0.0000 | |
| GSE53624 | IFT122 | 55764 | 54091 | 0.6004 | 0.0000 | |
| GSE63941 | IFT122 | 55764 | 220744_s_at | 0.1416 | 0.7567 | |
| GSE77861 | IFT122 | 55764 | 216678_at | -0.0525 | 0.6079 | |
| GSE97050 | IFT122 | 55764 | A_23_P212447 | 0.2629 | 0.2561 | |
| SRP007169 | IFT122 | 55764 | RNAseq | 1.7356 | 0.0035 | |
| SRP008496 | IFT122 | 55764 | RNAseq | 1.8102 | 0.0000 | |
| SRP064894 | IFT122 | 55764 | RNAseq | 0.7119 | 0.0002 | |
| SRP133303 | IFT122 | 55764 | RNAseq | 0.1008 | 0.4977 | |
| SRP159526 | IFT122 | 55764 | RNAseq | 0.8203 | 0.0588 | |
| SRP193095 | IFT122 | 55764 | RNAseq | 0.4391 | 0.0006 | |
| SRP219564 | IFT122 | 55764 | RNAseq | 0.5628 | 0.0513 | |
| TCGA | IFT122 | 55764 | RNAseq | -0.0386 | 0.5327 |
Upregulated datasets: 2; Downregulated datasets: 0.
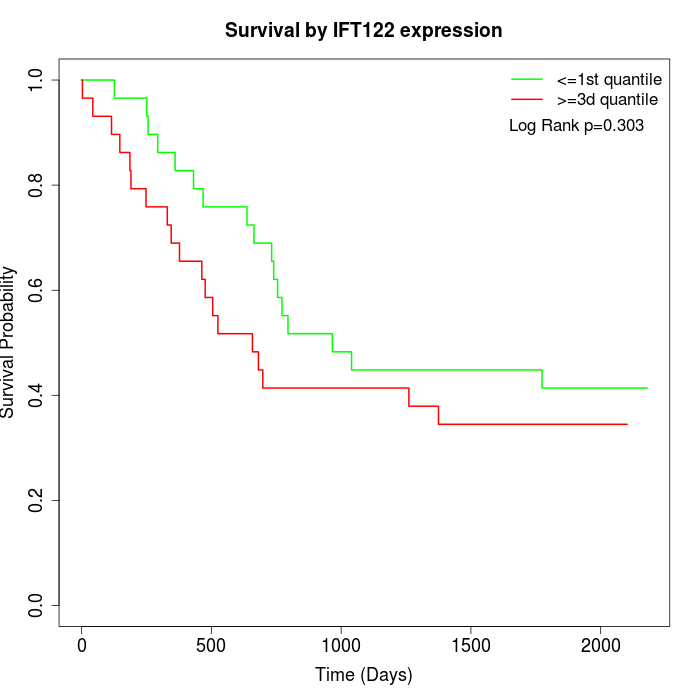
Survival by IFT122 expression:
Note: Click image to view full size file.
Copy number change of IFT122:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | IFT122 | 55764 | 17 | 0 | 13 | |
| GSE20123 | IFT122 | 55764 | 17 | 0 | 13 | |
| GSE43470 | IFT122 | 55764 | 19 | 0 | 24 | |
| GSE46452 | IFT122 | 55764 | 14 | 5 | 40 | |
| GSE47630 | IFT122 | 55764 | 18 | 3 | 19 | |
| GSE54993 | IFT122 | 55764 | 2 | 8 | 60 | |
| GSE54994 | IFT122 | 55764 | 32 | 3 | 18 | |
| GSE60625 | IFT122 | 55764 | 0 | 6 | 5 | |
| GSE74703 | IFT122 | 55764 | 17 | 0 | 19 | |
| GSE74704 | IFT122 | 55764 | 13 | 0 | 7 | |
| TCGA | IFT122 | 55764 | 58 | 5 | 33 |
Total number of gains: 207; Total number of losses: 30; Total Number of normals: 251.
Somatic mutations of IFT122:
Generating mutation plots.
Highly correlated genes for IFT122:
Showing top 20/359 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| IFT122 | GNG3 | 0.726697 | 4 | 0 | 4 |
| IFT122 | AATK | 0.722745 | 3 | 0 | 3 |
| IFT122 | SERPINA4 | 0.721662 | 4 | 0 | 4 |
| IFT122 | ZMAT4 | 0.718747 | 5 | 0 | 4 |
| IFT122 | NAT2 | 0.717632 | 3 | 0 | 3 |
| IFT122 | TSHR | 0.716052 | 4 | 0 | 4 |
| IFT122 | SLC17A7 | 0.715997 | 3 | 0 | 3 |
| IFT122 | CYP19A1 | 0.713252 | 3 | 0 | 3 |
| IFT122 | CNTD2 | 0.709501 | 4 | 0 | 4 |
| IFT122 | CYP11B1 | 0.708856 | 3 | 0 | 3 |
| IFT122 | REG3A | 0.708618 | 4 | 0 | 3 |
| IFT122 | TTR | 0.703173 | 4 | 0 | 4 |
| IFT122 | CNTN2 | 0.701613 | 4 | 0 | 4 |
| IFT122 | CA8 | 0.698787 | 3 | 0 | 3 |
| IFT122 | TGM4 | 0.695128 | 4 | 0 | 4 |
| IFT122 | ARHGEF38 | 0.694194 | 4 | 0 | 3 |
| IFT122 | KCNA1 | 0.693797 | 4 | 0 | 4 |
| IFT122 | OR2J2 | 0.692881 | 3 | 0 | 3 |
| IFT122 | CRP | 0.690116 | 4 | 0 | 4 |
| IFT122 | CACNG1 | 0.688461 | 3 | 0 | 3 |
For details and further investigation, click here