| Full name: cadherin 15 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 16q24.3 | ||
| Entrez ID: 1013 | HGNC ID: HGNC:1754 | Ensembl Gene: ENSG00000129910 | OMIM ID: 114019 |
| Drug and gene relationship at DGIdb | |||
Expression of CDH15:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CDH15 | 1013 | 206327_s_at | 0.0008 | 0.9982 | |
| GSE20347 | CDH15 | 1013 | 206327_s_at | 0.1151 | 0.2287 | |
| GSE23400 | CDH15 | 1013 | 206327_s_at | -0.2368 | 0.0000 | |
| GSE26886 | CDH15 | 1013 | 206328_at | 0.5221 | 0.0004 | |
| GSE29001 | CDH15 | 1013 | 206327_s_at | -0.0154 | 0.9473 | |
| GSE38129 | CDH15 | 1013 | 206327_s_at | -0.0608 | 0.4482 | |
| GSE45670 | CDH15 | 1013 | 206327_s_at | 0.1272 | 0.0822 | |
| GSE53622 | CDH15 | 1013 | 48855 | 0.5773 | 0.0003 | |
| GSE53624 | CDH15 | 1013 | 48855 | 1.4353 | 0.0000 | |
| GSE63941 | CDH15 | 1013 | 206327_s_at | 0.5199 | 0.0022 | |
| GSE77861 | CDH15 | 1013 | 206328_at | 0.0722 | 0.6694 | |
| TCGA | CDH15 | 1013 | RNAseq | -0.6590 | 0.3067 |
Upregulated datasets: 1; Downregulated datasets: 0.
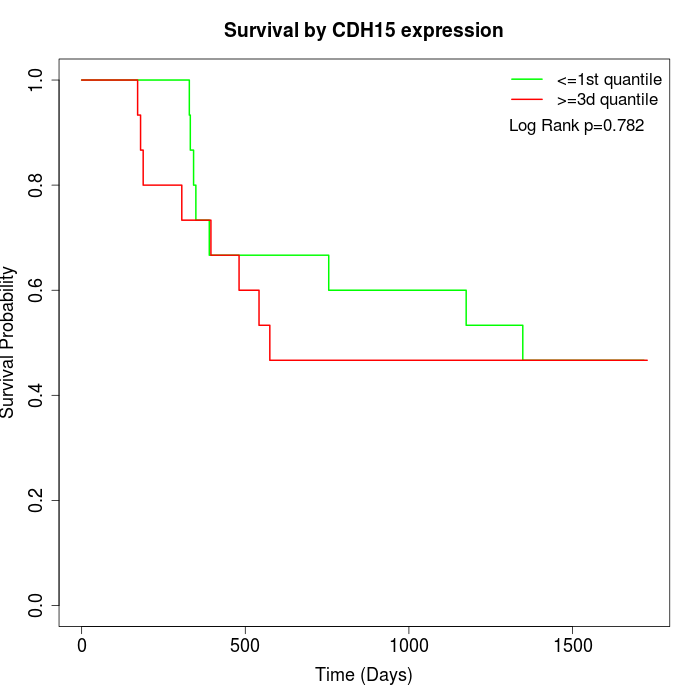
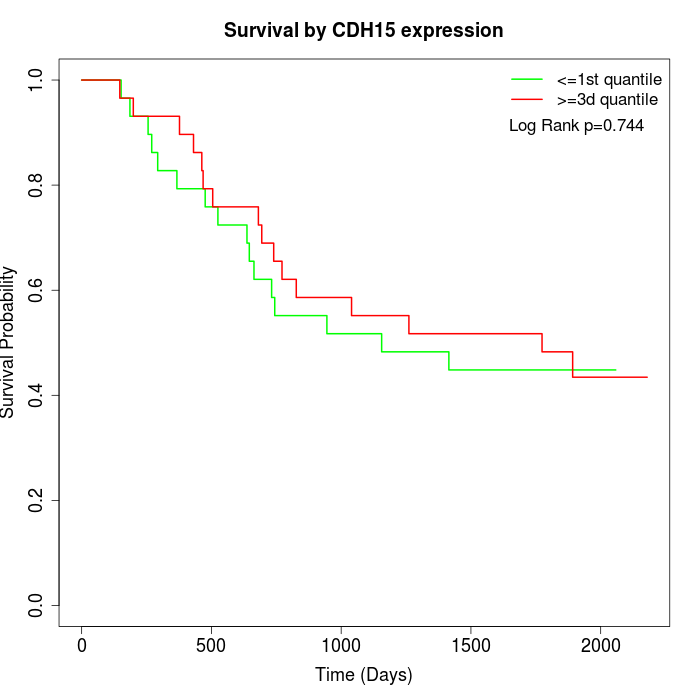
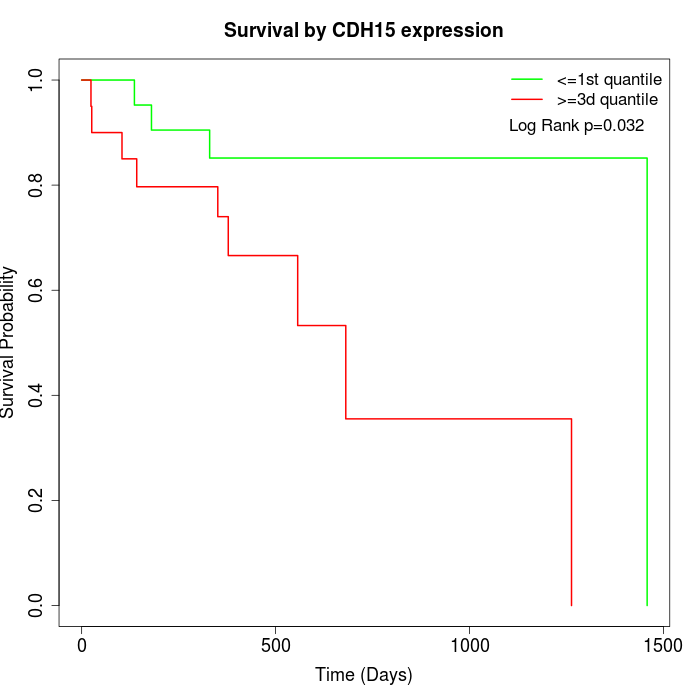
Survival by CDH15 expression:
Note: Click image to view full size file.
Copy number change of CDH15:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CDH15 | 1013 | 4 | 3 | 23 | |
| GSE20123 | CDH15 | 1013 | 4 | 3 | 23 | |
| GSE43470 | CDH15 | 1013 | 1 | 13 | 29 | |
| GSE46452 | CDH15 | 1013 | 38 | 1 | 20 | |
| GSE47630 | CDH15 | 1013 | 11 | 9 | 20 | |
| GSE54993 | CDH15 | 1013 | 3 | 4 | 63 | |
| GSE54994 | CDH15 | 1013 | 9 | 11 | 33 | |
| GSE60625 | CDH15 | 1013 | 4 | 0 | 7 | |
| GSE74703 | CDH15 | 1013 | 1 | 9 | 26 | |
| GSE74704 | CDH15 | 1013 | 3 | 2 | 15 | |
| TCGA | CDH15 | 1013 | 28 | 15 | 53 |
Total number of gains: 106; Total number of losses: 70; Total Number of normals: 312.
Somatic mutations of CDH15:
Generating mutation plots.
Highly correlated genes for CDH15:
Showing top 20/1114 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CDH15 | ZNF358 | 0.78488 | 3 | 0 | 3 |
| CDH15 | NOS2 | 0.741647 | 3 | 0 | 3 |
| CDH15 | UBE2D4 | 0.729069 | 3 | 0 | 3 |
| CDH15 | IL17F | 0.72594 | 3 | 0 | 3 |
| CDH15 | PSD | 0.720322 | 4 | 0 | 4 |
| CDH15 | RBP4 | 0.716972 | 3 | 0 | 3 |
| CDH15 | KCNQ2 | 0.716143 | 5 | 0 | 5 |
| CDH15 | LZTS1 | 0.711577 | 6 | 0 | 6 |
| CDH15 | PAPPA2 | 0.707171 | 5 | 0 | 5 |
| CDH15 | CXCR1 | 0.697772 | 5 | 0 | 5 |
| CDH15 | KLF16 | 0.694395 | 4 | 0 | 4 |
| CDH15 | SNAPC2 | 0.692205 | 4 | 0 | 4 |
| CDH15 | CHST4 | 0.692036 | 4 | 0 | 4 |
| CDH15 | KCNA1 | 0.691914 | 5 | 0 | 5 |
| CDH15 | ZNF580 | 0.690688 | 3 | 0 | 3 |
| CDH15 | CCDC106 | 0.689461 | 3 | 0 | 3 |
| CDH15 | LTB4R2 | 0.685238 | 5 | 0 | 4 |
| CDH15 | BCAT2 | 0.684554 | 3 | 0 | 3 |
| CDH15 | GPA33 | 0.684545 | 5 | 0 | 5 |
| CDH15 | CACNA1H | 0.681458 | 4 | 0 | 3 |
For details and further investigation, click here