| Full name: potassium voltage-gated channel subfamily D member 1 | Alias Symbol: Kv4.1 | ||
| Type: protein-coding gene | Cytoband: Xp11.23 | ||
| Entrez ID: 3750 | HGNC ID: HGNC:6237 | Ensembl Gene: ENSG00000102057 | OMIM ID: 300281 |
| Drug and gene relationship at DGIdb | |||
Expression of KCND1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCND1 | 3750 | 206842_at | -0.1313 | 0.6898 | |
| GSE20347 | KCND1 | 3750 | 206842_at | -0.0022 | 0.9822 | |
| GSE23400 | KCND1 | 3750 | 206842_at | -0.1888 | 0.0000 | |
| GSE26886 | KCND1 | 3750 | 206842_at | 0.2925 | 0.0956 | |
| GSE29001 | KCND1 | 3750 | 206842_at | -0.2811 | 0.1521 | |
| GSE38129 | KCND1 | 3750 | 206842_at | 0.0524 | 0.5231 | |
| GSE45670 | KCND1 | 3750 | 206842_at | -0.0765 | 0.4951 | |
| GSE53622 | KCND1 | 3750 | 44672 | 0.2681 | 0.1521 | |
| GSE53624 | KCND1 | 3750 | 44672 | 0.3582 | 0.0223 | |
| GSE63941 | KCND1 | 3750 | 206842_at | -0.1412 | 0.4991 | |
| GSE77861 | KCND1 | 3750 | 206842_at | -0.2299 | 0.1182 | |
| GSE97050 | KCND1 | 3750 | A_23_P315772 | 0.3514 | 0.1657 | |
| SRP064894 | KCND1 | 3750 | RNAseq | 1.0508 | 0.0000 | |
| SRP133303 | KCND1 | 3750 | RNAseq | 0.4709 | 0.1363 | |
| SRP159526 | KCND1 | 3750 | RNAseq | 1.1003 | 0.0079 | |
| SRP219564 | KCND1 | 3750 | RNAseq | 0.7159 | 0.0499 | |
| TCGA | KCND1 | 3750 | RNAseq | 0.0201 | 0.9038 |
Upregulated datasets: 2; Downregulated datasets: 0.
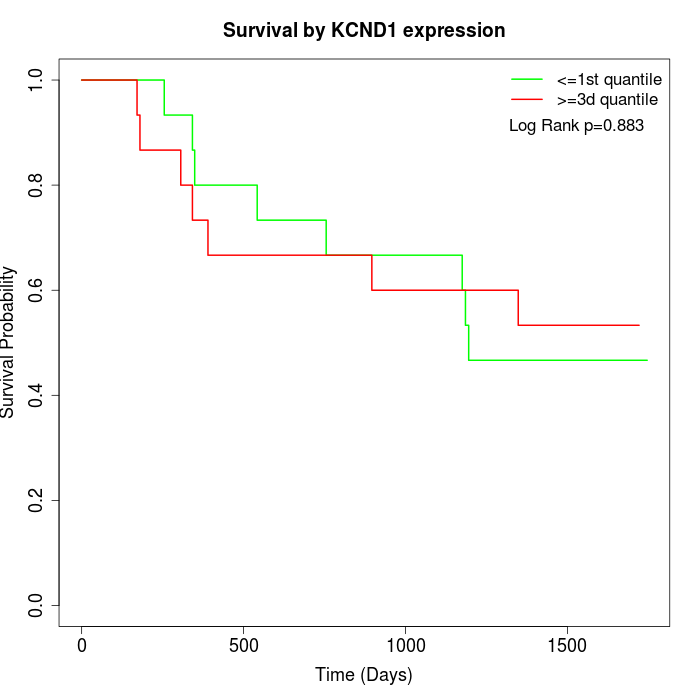
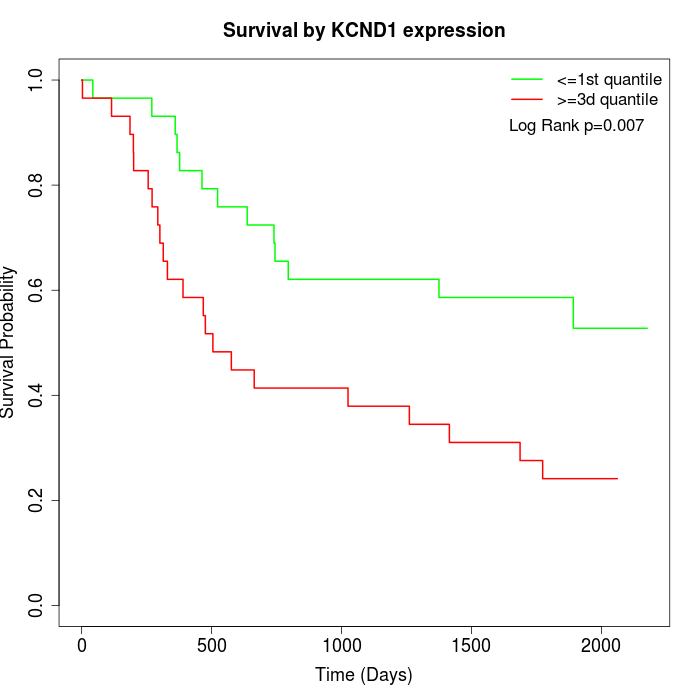
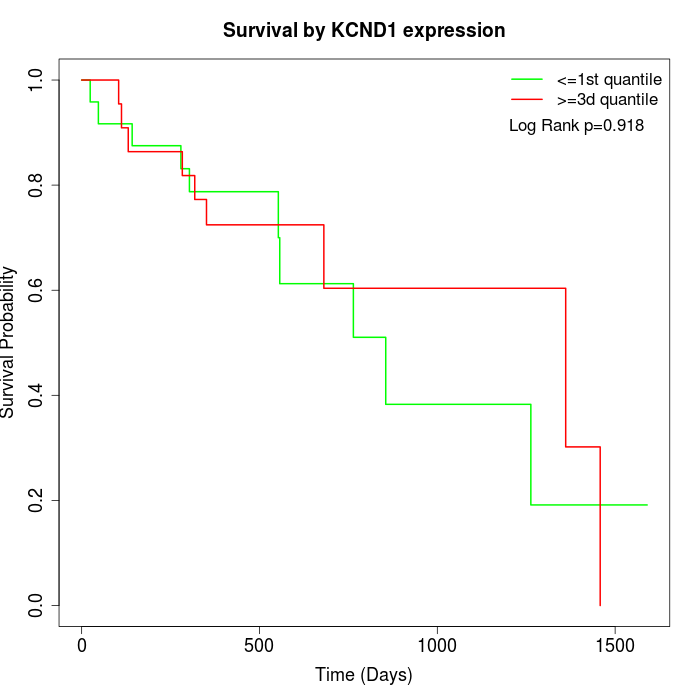
Survival by KCND1 expression:
Note: Click image to view full size file.
Copy number change of KCND1:
No record found for this gene.
Somatic mutations of KCND1:
Generating mutation plots.
Highly correlated genes for KCND1:
Showing top 20/337 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCND1 | CLDN2 | 0.797282 | 3 | 0 | 3 |
| KCND1 | SPIC | 0.786106 | 3 | 0 | 3 |
| KCND1 | NDN | 0.716767 | 3 | 0 | 3 |
| KCND1 | TMEM222 | 0.715815 | 3 | 0 | 3 |
| KCND1 | DNAH1 | 0.714043 | 4 | 0 | 3 |
| KCND1 | LMO2 | 0.709538 | 3 | 0 | 3 |
| KCND1 | OS9 | 0.702945 | 3 | 0 | 3 |
| KCND1 | YPEL2 | 0.690897 | 3 | 0 | 3 |
| KCND1 | BACH2 | 0.690734 | 5 | 0 | 5 |
| KCND1 | COL14A1 | 0.690084 | 4 | 0 | 4 |
| KCND1 | FOLR2 | 0.688339 | 3 | 0 | 3 |
| KCND1 | ZNF304 | 0.688191 | 3 | 0 | 3 |
| KCND1 | PIM2 | 0.681427 | 3 | 0 | 3 |
| KCND1 | SERPINA12 | 0.679925 | 3 | 0 | 3 |
| KCND1 | TRIM3 | 0.677322 | 4 | 0 | 3 |
| KCND1 | LDOC1 | 0.676458 | 3 | 0 | 3 |
| KCND1 | TMEM167B | 0.676179 | 3 | 0 | 3 |
| KCND1 | TMEM86B | 0.674904 | 3 | 0 | 3 |
| KCND1 | ZNF358 | 0.674828 | 5 | 0 | 5 |
| KCND1 | GNG7 | 0.674638 | 3 | 0 | 3 |
For details and further investigation, click here