| Full name: Spi-C transcription factor | Alias Symbol: MGC40611|SPI-C | ||
| Type: protein-coding gene | Cytoband: 12q23.2 | ||
| Entrez ID: 121599 | HGNC ID: HGNC:29549 | Ensembl Gene: ENSG00000166211 | OMIM ID: 612568 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of SPIC:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SPIC | 121599 | 1553851_at | 0.0726 | 0.7398 | |
| GSE26886 | SPIC | 121599 | 1553851_at | 0.2979 | 0.1122 | |
| GSE45670 | SPIC | 121599 | 1553851_at | 0.0851 | 0.2885 | |
| GSE53622 | SPIC | 121599 | 120309 | -0.2000 | 0.3759 | |
| GSE53624 | SPIC | 121599 | 120309 | -0.5172 | 0.0050 | |
| GSE63941 | SPIC | 121599 | 1553851_at | 0.0690 | 0.6111 | |
| GSE77861 | SPIC | 121599 | 1553851_at | -0.0641 | 0.3051 | |
| GSE97050 | SPIC | 121599 | A_32_P148275 | 0.4625 | 0.2462 | |
| TCGA | SPIC | 121599 | RNAseq | 1.1588 | 0.3396 |
Upregulated datasets: 0; Downregulated datasets: 0.
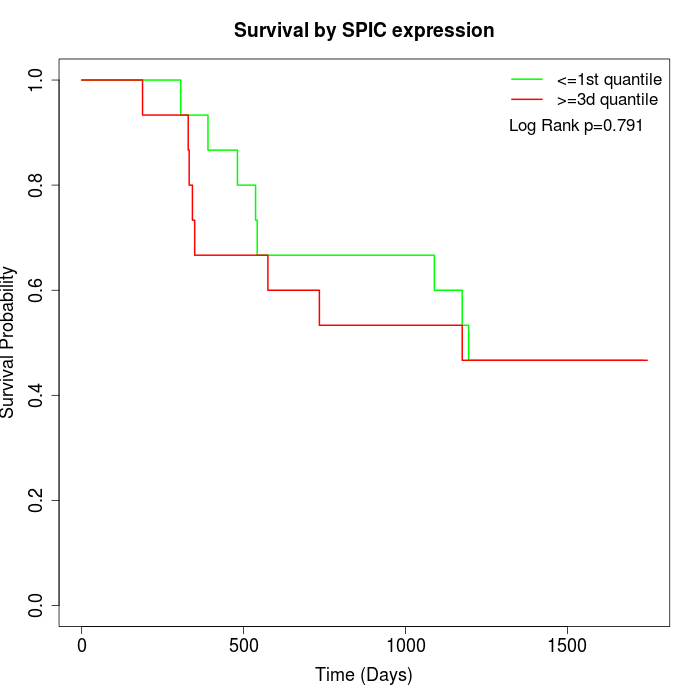
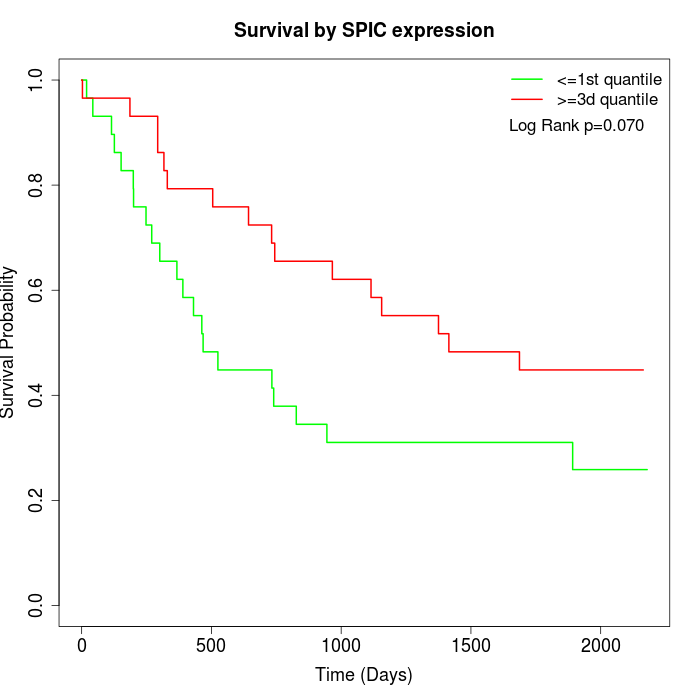
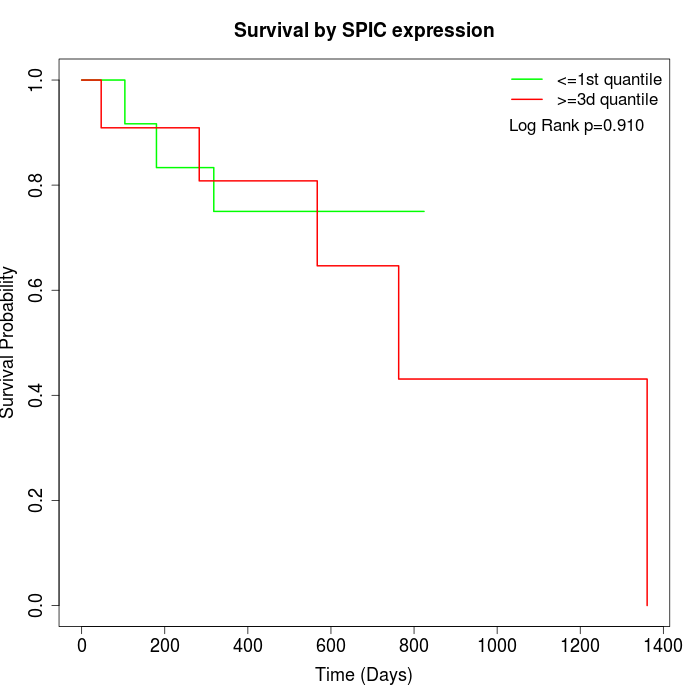
Survival by SPIC expression:
Note: Click image to view full size file.
Copy number change of SPIC:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SPIC | 121599 | 2 | 2 | 26 | |
| GSE20123 | SPIC | 121599 | 2 | 2 | 26 | |
| GSE43470 | SPIC | 121599 | 2 | 0 | 41 | |
| GSE46452 | SPIC | 121599 | 9 | 1 | 49 | |
| GSE47630 | SPIC | 121599 | 9 | 1 | 30 | |
| GSE54993 | SPIC | 121599 | 0 | 5 | 65 | |
| GSE54994 | SPIC | 121599 | 4 | 2 | 47 | |
| GSE60625 | SPIC | 121599 | 0 | 0 | 11 | |
| GSE74703 | SPIC | 121599 | 2 | 0 | 34 | |
| GSE74704 | SPIC | 121599 | 1 | 2 | 17 | |
| TCGA | SPIC | 121599 | 19 | 10 | 67 |
Total number of gains: 50; Total number of losses: 25; Total Number of normals: 413.
Somatic mutations of SPIC:
Generating mutation plots.
Highly correlated genes for SPIC:
Showing top 20/87 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SPIC | KCND1 | 0.786106 | 3 | 0 | 3 |
| SPIC | GAL | 0.774771 | 3 | 0 | 3 |
| SPIC | CEP250 | 0.773391 | 3 | 0 | 3 |
| SPIC | SCML4 | 0.768704 | 3 | 0 | 3 |
| SPIC | MRPL1 | 0.759752 | 3 | 0 | 3 |
| SPIC | PLAG1 | 0.758486 | 3 | 0 | 3 |
| SPIC | ATG13 | 0.738302 | 3 | 0 | 3 |
| SPIC | EHMT2 | 0.735498 | 3 | 0 | 3 |
| SPIC | RCHY1 | 0.733697 | 3 | 0 | 3 |
| SPIC | IRX3 | 0.732736 | 3 | 0 | 3 |
| SPIC | MLLT1 | 0.729434 | 3 | 0 | 3 |
| SPIC | POLR2K | 0.728898 | 3 | 0 | 3 |
| SPIC | APEX1 | 0.727179 | 3 | 0 | 3 |
| SPIC | GATA3 | 0.724049 | 3 | 0 | 3 |
| SPIC | CNIH3 | 0.722642 | 3 | 0 | 3 |
| SPIC | ZNF432 | 0.719913 | 3 | 0 | 3 |
| SPIC | TBC1D23 | 0.719575 | 3 | 0 | 3 |
| SPIC | FNBP1L | 0.713952 | 3 | 0 | 3 |
| SPIC | ZNF581 | 0.712963 | 3 | 0 | 3 |
| SPIC | IL18BP | 0.709671 | 3 | 0 | 3 |
For details and further investigation, click here