| Full name: LIM domain only 2 | Alias Symbol: TTG2|RHOM2|RBTN2 | ||
| Type: protein-coding gene | Cytoband: 11p13 | ||
| Entrez ID: 4005 | HGNC ID: HGNC:6642 | Ensembl Gene: ENSG00000135363 | OMIM ID: 180385 |
| Drug and gene relationship at DGIdb | |||
Expression of LMO2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LMO2 | 4005 | 204249_s_at | -0.7024 | 0.1514 | |
| GSE20347 | LMO2 | 4005 | 204249_s_at | -1.3475 | 0.0000 | |
| GSE23400 | LMO2 | 4005 | 204249_s_at | -1.0964 | 0.0000 | |
| GSE26886 | LMO2 | 4005 | 204249_s_at | -2.8467 | 0.0000 | |
| GSE29001 | LMO2 | 4005 | 204249_s_at | -1.1818 | 0.0044 | |
| GSE38129 | LMO2 | 4005 | 204249_s_at | -1.1371 | 0.0000 | |
| GSE45670 | LMO2 | 4005 | 204249_s_at | -0.4444 | 0.1664 | |
| GSE53622 | LMO2 | 4005 | 104035 | -1.1228 | 0.0000 | |
| GSE53624 | LMO2 | 4005 | 104035 | -1.5044 | 0.0000 | |
| GSE63941 | LMO2 | 4005 | 204249_s_at | -0.9866 | 0.0465 | |
| GSE77861 | LMO2 | 4005 | 204249_s_at | -0.7722 | 0.0153 | |
| GSE97050 | LMO2 | 4005 | A_23_P53126 | -0.1041 | 0.8056 | |
| SRP007169 | LMO2 | 4005 | RNAseq | -1.7312 | 0.0068 | |
| SRP064894 | LMO2 | 4005 | RNAseq | -0.8782 | 0.0034 | |
| SRP133303 | LMO2 | 4005 | RNAseq | -1.3377 | 0.0000 | |
| SRP159526 | LMO2 | 4005 | RNAseq | -1.5878 | 0.0001 | |
| SRP193095 | LMO2 | 4005 | RNAseq | -1.3615 | 0.0000 | |
| SRP219564 | LMO2 | 4005 | RNAseq | -0.8159 | 0.0394 | |
| TCGA | LMO2 | 4005 | RNAseq | -0.1860 | 0.1043 |
Upregulated datasets: 0; Downregulated datasets: 11.
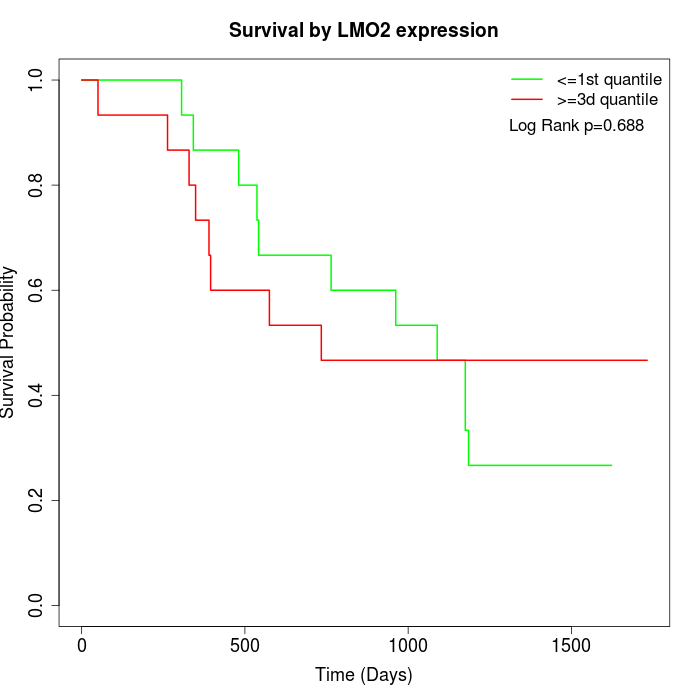
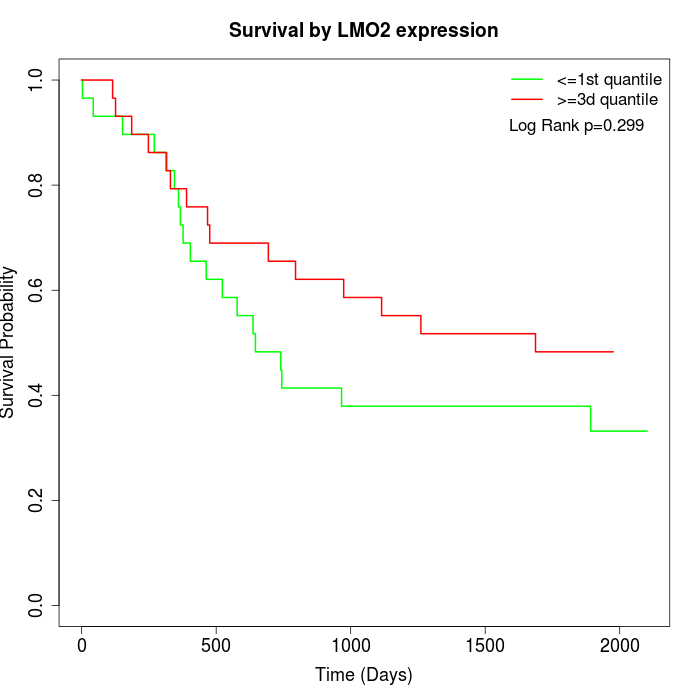
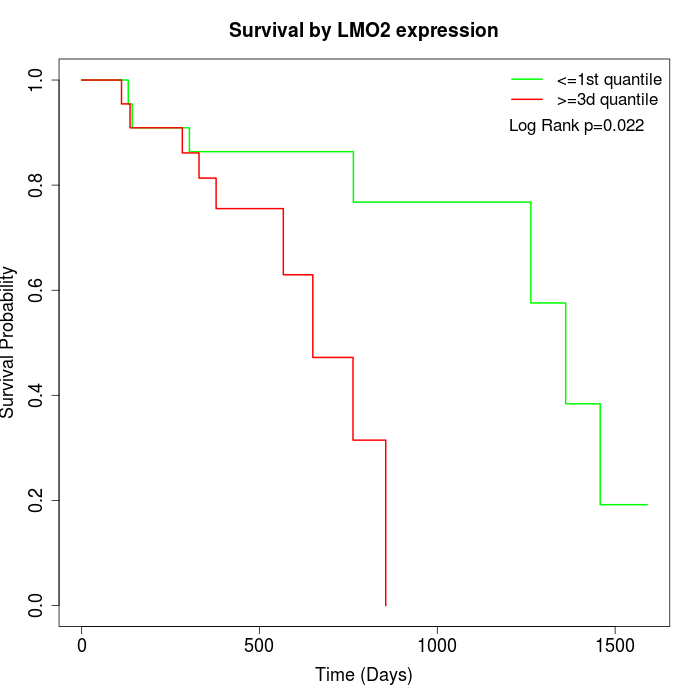
Survival by LMO2 expression:
Note: Click image to view full size file.
Copy number change of LMO2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LMO2 | 4005 | 1 | 6 | 23 | |
| GSE20123 | LMO2 | 4005 | 1 | 5 | 24 | |
| GSE43470 | LMO2 | 4005 | 2 | 4 | 37 | |
| GSE46452 | LMO2 | 4005 | 7 | 5 | 47 | |
| GSE47630 | LMO2 | 4005 | 3 | 10 | 27 | |
| GSE54993 | LMO2 | 4005 | 3 | 1 | 66 | |
| GSE54994 | LMO2 | 4005 | 4 | 9 | 40 | |
| GSE60625 | LMO2 | 4005 | 0 | 0 | 11 | |
| GSE74703 | LMO2 | 4005 | 2 | 2 | 32 | |
| GSE74704 | LMO2 | 4005 | 0 | 3 | 17 | |
| TCGA | LMO2 | 4005 | 13 | 24 | 59 |
Total number of gains: 36; Total number of losses: 69; Total Number of normals: 383.
Somatic mutations of LMO2:
Generating mutation plots.
Highly correlated genes for LMO2:
Showing top 20/1530 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LMO2 | EVC2 | 0.776554 | 3 | 0 | 3 |
| LMO2 | ZACN | 0.741068 | 3 | 0 | 3 |
| LMO2 | SPAG17 | 0.736925 | 4 | 0 | 3 |
| LMO2 | IMPACT | 0.733535 | 3 | 0 | 3 |
| LMO2 | RBM20 | 0.730706 | 4 | 0 | 4 |
| LMO2 | CHCHD1 | 0.729082 | 4 | 0 | 4 |
| LMO2 | SEC23IP | 0.726894 | 3 | 0 | 3 |
| LMO2 | MPP7 | 0.724404 | 7 | 0 | 7 |
| LMO2 | TMEM234 | 0.72259 | 3 | 0 | 3 |
| LMO2 | TMPRSS11B | 0.721189 | 6 | 0 | 6 |
| LMO2 | UBQLN1 | 0.72112 | 3 | 0 | 3 |
| LMO2 | EPB41L3 | 0.72067 | 12 | 0 | 12 |
| LMO2 | KAT2B | 0.719063 | 11 | 0 | 11 |
| LMO2 | CLEC3B | 0.718393 | 3 | 0 | 3 |
| LMO2 | CRISP3 | 0.713161 | 10 | 0 | 10 |
| LMO2 | NAGA | 0.712191 | 4 | 0 | 3 |
| LMO2 | FAM214A | 0.711162 | 6 | 0 | 6 |
| LMO2 | FCER1A | 0.709689 | 10 | 0 | 10 |
| LMO2 | KCND1 | 0.709538 | 3 | 0 | 3 |
| LMO2 | TIFA | 0.707507 | 7 | 0 | 7 |
For details and further investigation, click here