| Full name: kelch like family member 10 | Alias Symbol: FLJ32662 | ||
| Type: protein-coding gene | Cytoband: 17q21.2 | ||
| Entrez ID: 317719 | HGNC ID: HGNC:18829 | Ensembl Gene: ENSG00000161594 | OMIM ID: 608778 |
| Drug and gene relationship at DGIdb | |||
Expression of KLHL10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KLHL10 | 317719 | 1552425_a_at | -0.5483 | 0.1438 | |
| GSE26886 | KLHL10 | 317719 | 1552425_a_at | -0.2796 | 0.0193 | |
| GSE45670 | KLHL10 | 317719 | 1552425_a_at | -0.2172 | 0.0188 | |
| GSE53622 | KLHL10 | 317719 | 145718 | -1.0354 | 0.0000 | |
| GSE53624 | KLHL10 | 317719 | 145718 | -1.2304 | 0.0000 | |
| GSE63941 | KLHL10 | 317719 | 1552425_a_at | -0.1199 | 0.3595 | |
| GSE77861 | KLHL10 | 317719 | 1552425_a_at | -0.1347 | 0.0860 | |
| GSE97050 | KLHL10 | 317719 | A_23_P27128 | -0.4051 | 0.2071 | |
| SRP219564 | KLHL10 | 317719 | RNAseq | 0.0136 | 0.9806 | |
| TCGA | KLHL10 | 317719 | RNAseq | -1.7211 | 0.0078 |
Upregulated datasets: 0; Downregulated datasets: 3.
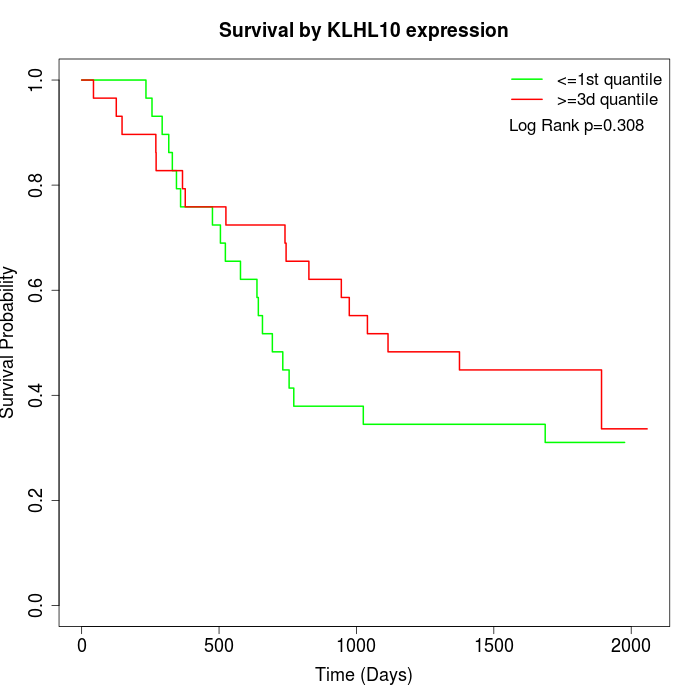
Survival by KLHL10 expression:
Note: Click image to view full size file.
Copy number change of KLHL10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KLHL10 | 317719 | 6 | 2 | 22 | |
| GSE20123 | KLHL10 | 317719 | 6 | 2 | 22 | |
| GSE43470 | KLHL10 | 317719 | 1 | 2 | 40 | |
| GSE46452 | KLHL10 | 317719 | 34 | 0 | 25 | |
| GSE47630 | KLHL10 | 317719 | 8 | 1 | 31 | |
| GSE54993 | KLHL10 | 317719 | 3 | 4 | 63 | |
| GSE54994 | KLHL10 | 317719 | 8 | 5 | 40 | |
| GSE60625 | KLHL10 | 317719 | 4 | 0 | 7 | |
| GSE74703 | KLHL10 | 317719 | 1 | 1 | 34 | |
| GSE74704 | KLHL10 | 317719 | 4 | 1 | 15 | |
| TCGA | KLHL10 | 317719 | 23 | 7 | 66 |
Total number of gains: 98; Total number of losses: 25; Total Number of normals: 365.
Somatic mutations of KLHL10:
Generating mutation plots.
Highly correlated genes for KLHL10:
Showing top 20/525 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KLHL10 | OR10G8 | 0.87432 | 3 | 0 | 3 |
| KLHL10 | KRTAP20-1 | 0.834298 | 3 | 0 | 3 |
| KLHL10 | OR2Z1 | 0.833999 | 3 | 0 | 3 |
| KLHL10 | GADL1 | 0.811709 | 3 | 0 | 3 |
| KLHL10 | FAM163B | 0.810433 | 3 | 0 | 3 |
| KLHL10 | HRCT1 | 0.807584 | 4 | 0 | 4 |
| KLHL10 | C20orf173 | 0.806692 | 3 | 0 | 3 |
| KLHL10 | KCNF1 | 0.800386 | 3 | 0 | 3 |
| KLHL10 | MUC8 | 0.795388 | 3 | 0 | 3 |
| KLHL10 | GATA4 | 0.787015 | 3 | 0 | 3 |
| KLHL10 | BARHL2 | 0.775088 | 3 | 0 | 3 |
| KLHL10 | TEN1 | 0.767396 | 3 | 0 | 3 |
| KLHL10 | CPNE6 | 0.766079 | 4 | 0 | 3 |
| KLHL10 | PCDH1 | 0.758786 | 4 | 0 | 4 |
| KLHL10 | OPN4 | 0.756114 | 3 | 0 | 3 |
| KLHL10 | MYO1A | 0.753348 | 4 | 0 | 4 |
| KLHL10 | APOBEC1 | 0.751718 | 4 | 0 | 4 |
| KLHL10 | MUC22 | 0.750213 | 3 | 0 | 3 |
| KLHL10 | FAM71E2 | 0.746629 | 3 | 0 | 3 |
| KLHL10 | SH3GLB2 | 0.743925 | 3 | 0 | 3 |
For details and further investigation, click here