| Full name: tripartite motif family like 2 | Alias Symbol: FLJ25801|SPRYD6 | ||
| Type: protein-coding gene | Cytoband: 4q35.2 | ||
| Entrez ID: 205860 | HGNC ID: HGNC:26378 | Ensembl Gene: ENSG00000179046 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of TRIML2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIML2 | 205860 | 1552580_at | -0.0596 | 0.7840 | |
| GSE26886 | TRIML2 | 205860 | 1552580_at | 0.2382 | 0.0744 | |
| GSE45670 | TRIML2 | 205860 | 1552580_at | 0.1373 | 0.3604 | |
| GSE53622 | TRIML2 | 205860 | 4692 | 0.8434 | 0.0002 | |
| GSE53624 | TRIML2 | 205860 | 4692 | 1.8242 | 0.0000 | |
| GSE63941 | TRIML2 | 205860 | 1552580_at | 0.2187 | 0.3494 | |
| GSE77861 | TRIML2 | 205860 | 1552580_at | -0.3114 | 0.0805 | |
| SRP133303 | TRIML2 | 205860 | RNAseq | 0.1756 | 0.3347 | |
| SRP193095 | TRIML2 | 205860 | RNAseq | 0.1333 | 0.5746 | |
| TCGA | TRIML2 | 205860 | RNAseq | 4.5350 | 0.0000 |
Upregulated datasets: 2; Downregulated datasets: 0.
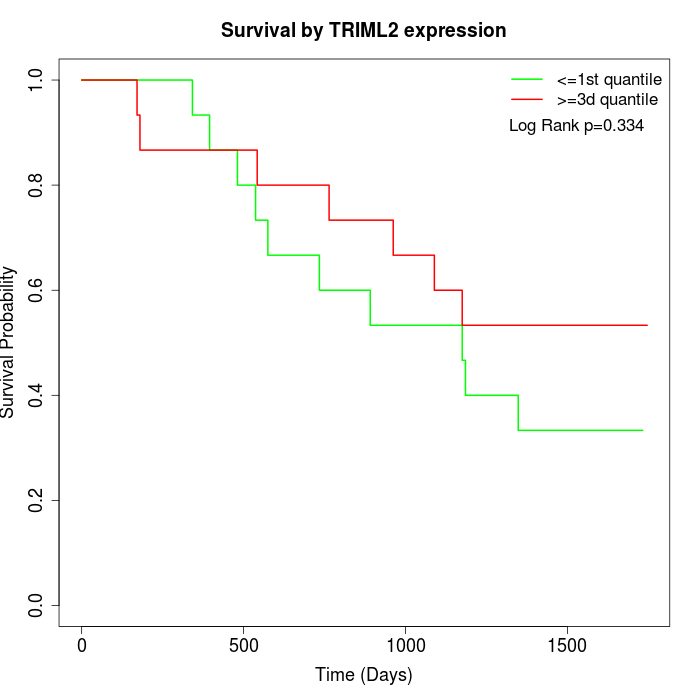
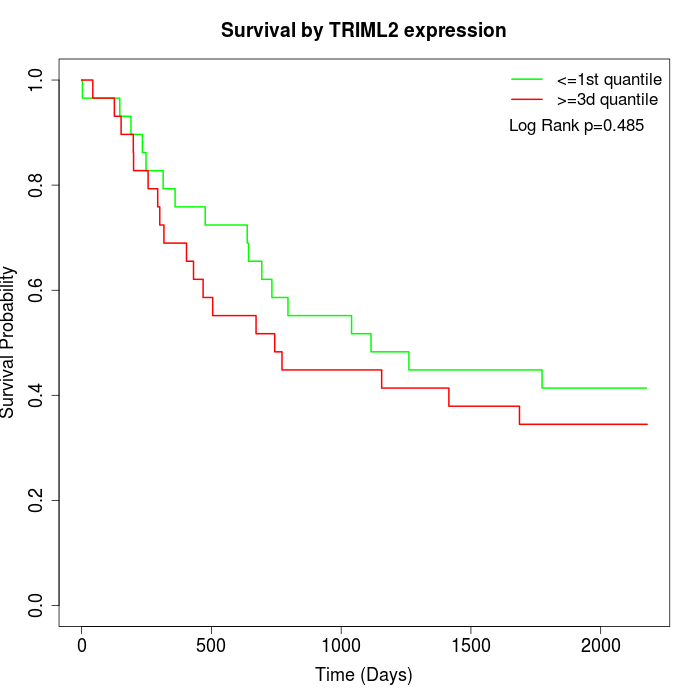
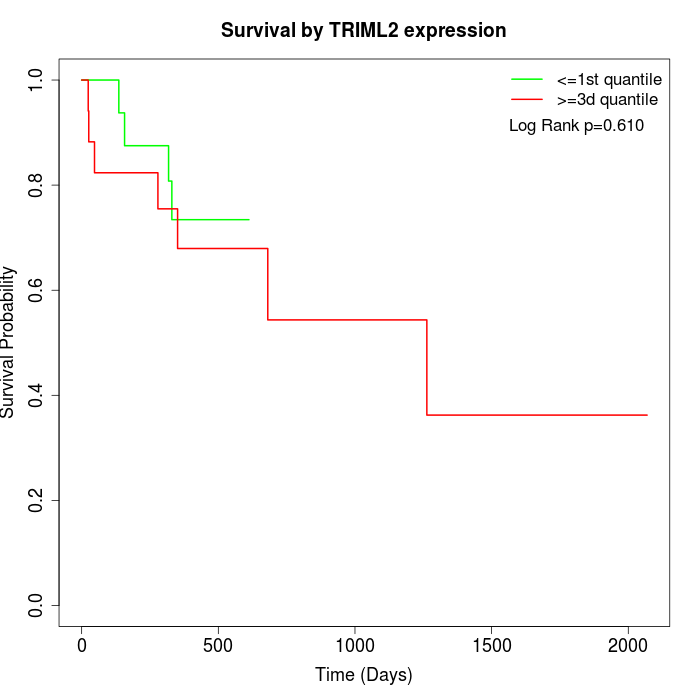
Survival by TRIML2 expression:
Note: Click image to view full size file.
Copy number change of TRIML2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIML2 | 205860 | 2 | 12 | 16 | |
| GSE20123 | TRIML2 | 205860 | 2 | 12 | 16 | |
| GSE43470 | TRIML2 | 205860 | 0 | 16 | 27 | |
| GSE46452 | TRIML2 | 205860 | 1 | 36 | 22 | |
| GSE47630 | TRIML2 | 205860 | 0 | 22 | 18 | |
| GSE54993 | TRIML2 | 205860 | 10 | 0 | 60 | |
| GSE54994 | TRIML2 | 205860 | 2 | 13 | 38 | |
| GSE60625 | TRIML2 | 205860 | 5 | 1 | 5 | |
| GSE74703 | TRIML2 | 205860 | 0 | 13 | 23 | |
| GSE74704 | TRIML2 | 205860 | 0 | 6 | 14 | |
| TCGA | TRIML2 | 205860 | 10 | 37 | 49 |
Total number of gains: 32; Total number of losses: 168; Total Number of normals: 288.
Somatic mutations of TRIML2:
Generating mutation plots.
Highly correlated genes for TRIML2:
Showing top 20/22 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIML2 | CHD5 | 0.75074 | 3 | 0 | 3 |
| TRIML2 | KCNF1 | 0.744397 | 3 | 0 | 3 |
| TRIML2 | GNG8 | 0.689828 | 3 | 0 | 3 |
| TRIML2 | REG3G | 0.685128 | 3 | 0 | 3 |
| TRIML2 | TENM1 | 0.65942 | 3 | 0 | 3 |
| TRIML2 | USP2 | 0.652157 | 3 | 0 | 3 |
| TRIML2 | HMCN2 | 0.645804 | 4 | 0 | 3 |
| TRIML2 | DENND2A | 0.645216 | 3 | 0 | 3 |
| TRIML2 | SARDH | 0.643142 | 3 | 0 | 3 |
| TRIML2 | NACAD | 0.63703 | 3 | 0 | 3 |
| TRIML2 | MPL | 0.636085 | 3 | 0 | 3 |
| TRIML2 | CACNA1H | 0.634883 | 3 | 0 | 3 |
| TRIML2 | COX6A2 | 0.597442 | 3 | 0 | 3 |
| TRIML2 | PNMT | 0.576763 | 3 | 0 | 3 |
| TRIML2 | DNALI1 | 0.563007 | 3 | 0 | 3 |
| TRIML2 | SLC8A2 | 0.560507 | 3 | 0 | 3 |
| TRIML2 | DNAH17-AS1 | 0.55691 | 3 | 0 | 3 |
| TRIML2 | REEP2 | 0.55008 | 3 | 0 | 3 |
| TRIML2 | KCNN3 | 0.549238 | 3 | 0 | 3 |
| TRIML2 | IL20 | 0.536673 | 3 | 0 | 3 |
For details and further investigation, click here