| Full name: potassium inwardly rectifying channel subfamily J member 2 | Alias Symbol: Kir2.1|IRK1|LQT7 | ||
| Type: protein-coding gene | Cytoband: 17q24.3 | ||
| Entrez ID: 3759 | HGNC ID: HGNC:6263 | Ensembl Gene: ENSG00000123700 | OMIM ID: 600681 |
| Drug and gene relationship at DGIdb | |||
KCNJ2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04921 | Oxytocin signaling pathway | |
| hsa04924 | Renin secretion | |
| hsa04971 | Gastric acid secretion |
Expression of KCNJ2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNJ2 | 3759 | 206765_at | -0.1898 | 0.9211 | |
| GSE20347 | KCNJ2 | 3759 | 206765_at | -0.8630 | 0.0116 | |
| GSE23400 | KCNJ2 | 3759 | 206765_at | -0.0462 | 0.7293 | |
| GSE26886 | KCNJ2 | 3759 | 206765_at | -1.1580 | 0.0423 | |
| GSE29001 | KCNJ2 | 3759 | 206765_at | -1.0276 | 0.0545 | |
| GSE38129 | KCNJ2 | 3759 | 206765_at | -0.5371 | 0.0927 | |
| GSE45670 | KCNJ2 | 3759 | 206765_at | 0.0557 | 0.8814 | |
| GSE53622 | KCNJ2 | 3759 | 15098 | -0.2541 | 0.1566 | |
| GSE53624 | KCNJ2 | 3759 | 15098 | -0.3953 | 0.0023 | |
| GSE63941 | KCNJ2 | 3759 | 206765_at | -4.6616 | 0.0009 | |
| GSE77861 | KCNJ2 | 3759 | 206765_at | -0.3051 | 0.4747 | |
| GSE97050 | KCNJ2 | 3759 | A_23_P329261 | 0.0050 | 0.9897 | |
| SRP007169 | KCNJ2 | 3759 | RNAseq | 0.4248 | 0.5283 | |
| SRP008496 | KCNJ2 | 3759 | RNAseq | -0.2492 | 0.5949 | |
| SRP064894 | KCNJ2 | 3759 | RNAseq | 0.1500 | 0.5644 | |
| SRP133303 | KCNJ2 | 3759 | RNAseq | 0.0279 | 0.9298 | |
| SRP159526 | KCNJ2 | 3759 | RNAseq | -0.4727 | 0.3353 | |
| SRP193095 | KCNJ2 | 3759 | RNAseq | -0.2774 | 0.3484 | |
| SRP219564 | KCNJ2 | 3759 | RNAseq | -0.4481 | 0.4113 | |
| TCGA | KCNJ2 | 3759 | RNAseq | 0.1574 | 0.3540 |
Upregulated datasets: 0; Downregulated datasets: 2.
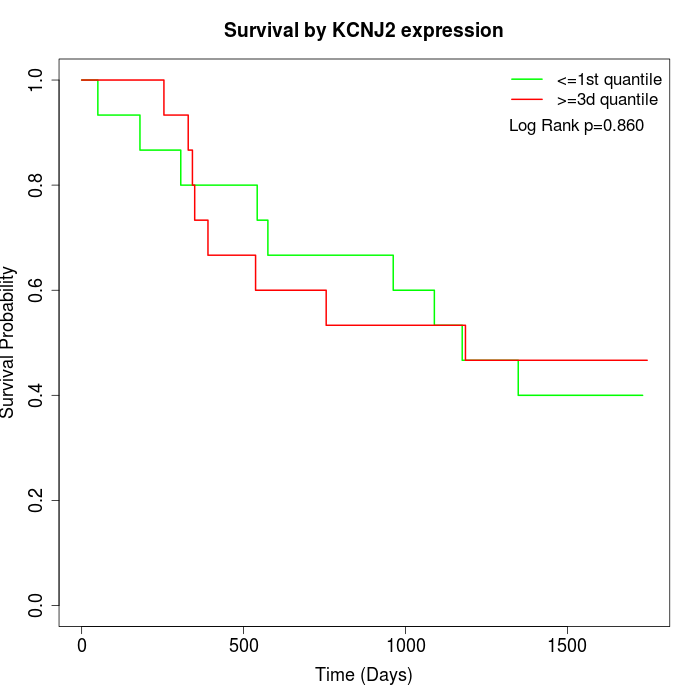
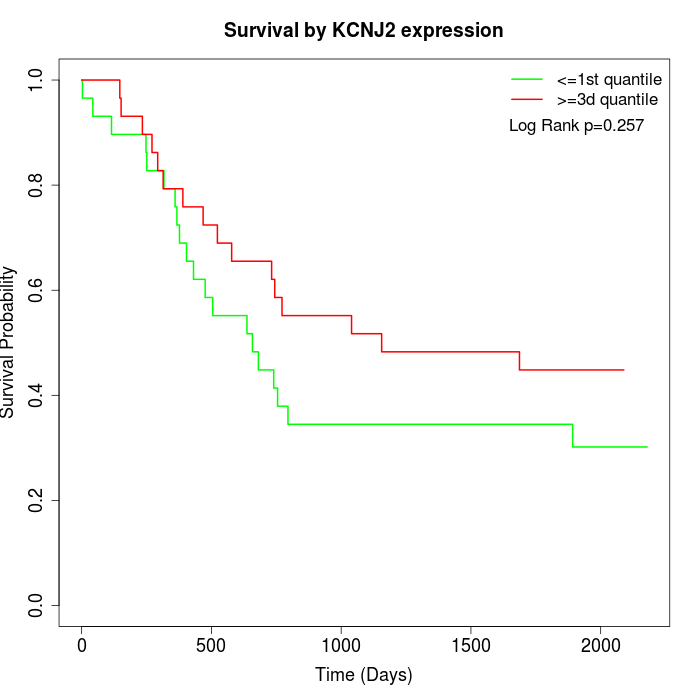
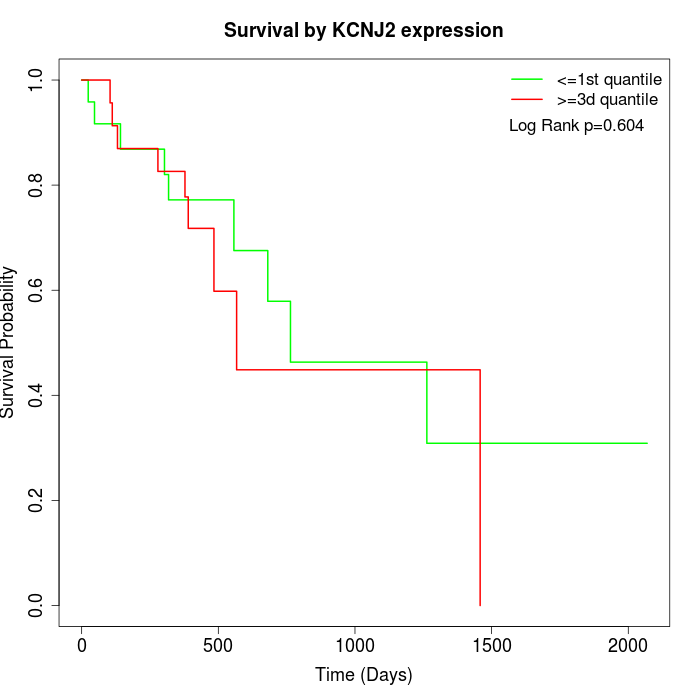
Survival by KCNJ2 expression:
Note: Click image to view full size file.
Copy number change of KCNJ2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNJ2 | 3759 | 3 | 1 | 26 | |
| GSE20123 | KCNJ2 | 3759 | 3 | 1 | 26 | |
| GSE43470 | KCNJ2 | 3759 | 7 | 1 | 35 | |
| GSE46452 | KCNJ2 | 3759 | 32 | 0 | 27 | |
| GSE47630 | KCNJ2 | 3759 | 7 | 1 | 32 | |
| GSE54993 | KCNJ2 | 3759 | 2 | 5 | 63 | |
| GSE54994 | KCNJ2 | 3759 | 9 | 4 | 40 | |
| GSE60625 | KCNJ2 | 3759 | 4 | 3 | 4 | |
| GSE74703 | KCNJ2 | 3759 | 7 | 0 | 29 | |
| GSE74704 | KCNJ2 | 3759 | 3 | 1 | 16 | |
| TCGA | KCNJ2 | 3759 | 31 | 10 | 55 |
Total number of gains: 108; Total number of losses: 27; Total Number of normals: 353.
Somatic mutations of KCNJ2:
Generating mutation plots.
Highly correlated genes for KCNJ2:
Showing top 20/283 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNJ2 | NAGA | 0.793674 | 3 | 0 | 3 |
| KCNJ2 | ANPEP | 0.763928 | 3 | 0 | 3 |
| KCNJ2 | SMC5 | 0.746353 | 3 | 0 | 3 |
| KCNJ2 | PRRX1 | 0.740336 | 3 | 0 | 3 |
| KCNJ2 | PDE7B | 0.732065 | 3 | 0 | 3 |
| KCNJ2 | ETS1 | 0.728234 | 3 | 0 | 3 |
| KCNJ2 | FAM71F2 | 0.727973 | 3 | 0 | 3 |
| KCNJ2 | ZNF441 | 0.717085 | 3 | 0 | 3 |
| KCNJ2 | RARS2 | 0.715975 | 4 | 0 | 4 |
| KCNJ2 | UNC5C | 0.71551 | 3 | 0 | 3 |
| KCNJ2 | DRAM2 | 0.71383 | 3 | 0 | 3 |
| KCNJ2 | RHOH | 0.710629 | 3 | 0 | 3 |
| KCNJ2 | FBXL3 | 0.708808 | 3 | 0 | 3 |
| KCNJ2 | CD59 | 0.707728 | 3 | 0 | 3 |
| KCNJ2 | B3GNT7 | 0.703926 | 3 | 0 | 3 |
| KCNJ2 | CXCL5 | 0.702784 | 3 | 0 | 3 |
| KCNJ2 | TMX4 | 0.698891 | 3 | 0 | 3 |
| KCNJ2 | ARG2 | 0.698737 | 3 | 0 | 3 |
| KCNJ2 | DPY19L3 | 0.698061 | 3 | 0 | 3 |
| KCNJ2 | ELMOD2 | 0.696564 | 3 | 0 | 3 |
For details and further investigation, click here