| Full name: DNA damage regulated autophagy modulator 2 | Alias Symbol: MGC54289|PRO180|WWFQ154|RP5-1180E21.1 | ||
| Type: protein-coding gene | Cytoband: 1p13.3 | ||
| Entrez ID: 128338 | HGNC ID: HGNC:28769 | Ensembl Gene: ENSG00000156171 | OMIM ID: 613360 |
| Drug and gene relationship at DGIdb | |||
Expression of DRAM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DRAM2 | 128338 | 225230_at | -0.2738 | 0.6448 | |
| GSE26886 | DRAM2 | 128338 | 225230_at | -0.5850 | 0.0510 | |
| GSE45670 | DRAM2 | 128338 | 225230_at | -0.2131 | 0.2538 | |
| GSE53622 | DRAM2 | 128338 | 22214 | -0.0190 | 0.8287 | |
| GSE53624 | DRAM2 | 128338 | 22214 | -0.0587 | 0.4407 | |
| GSE63941 | DRAM2 | 128338 | 225230_at | 0.0952 | 0.8685 | |
| GSE77861 | DRAM2 | 128338 | 225230_at | -0.2982 | 0.1740 | |
| GSE97050 | DRAM2 | 128338 | A_24_P268160 | 0.3784 | 0.2151 | |
| SRP007169 | DRAM2 | 128338 | RNAseq | 0.2282 | 0.5744 | |
| SRP008496 | DRAM2 | 128338 | RNAseq | 0.0427 | 0.8682 | |
| SRP064894 | DRAM2 | 128338 | RNAseq | 0.1413 | 0.4122 | |
| SRP133303 | DRAM2 | 128338 | RNAseq | -0.0641 | 0.7059 | |
| SRP159526 | DRAM2 | 128338 | RNAseq | -0.4404 | 0.1152 | |
| SRP193095 | DRAM2 | 128338 | RNAseq | -0.4753 | 0.0007 | |
| SRP219564 | DRAM2 | 128338 | RNAseq | -0.2097 | 0.3883 | |
| TCGA | DRAM2 | 128338 | RNAseq | -0.0770 | 0.1381 |
Upregulated datasets: 0; Downregulated datasets: 0.
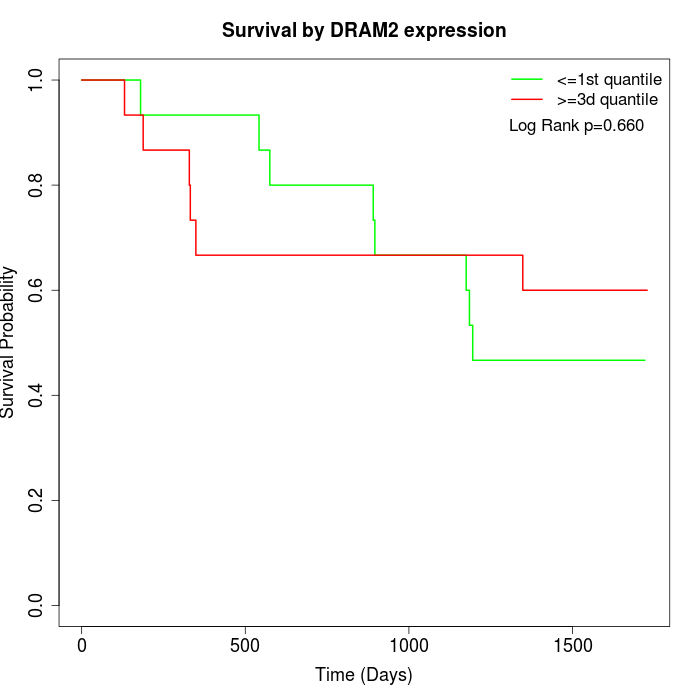
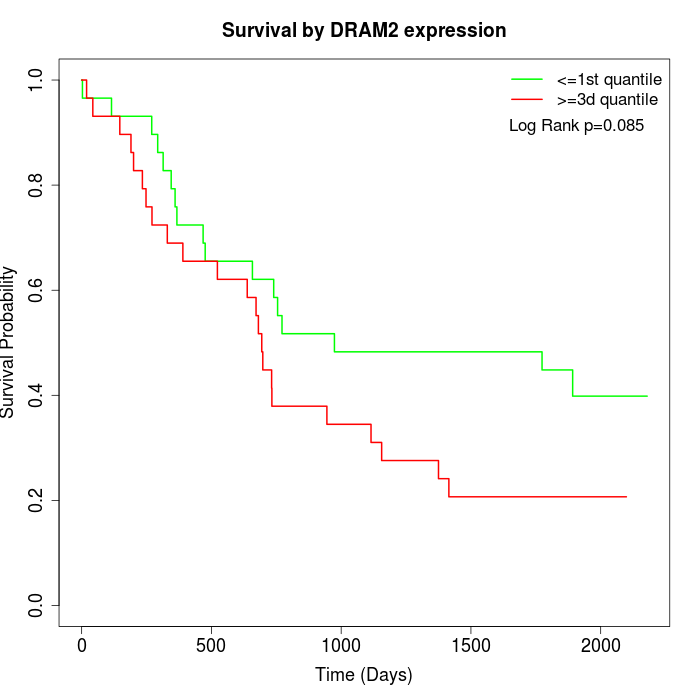
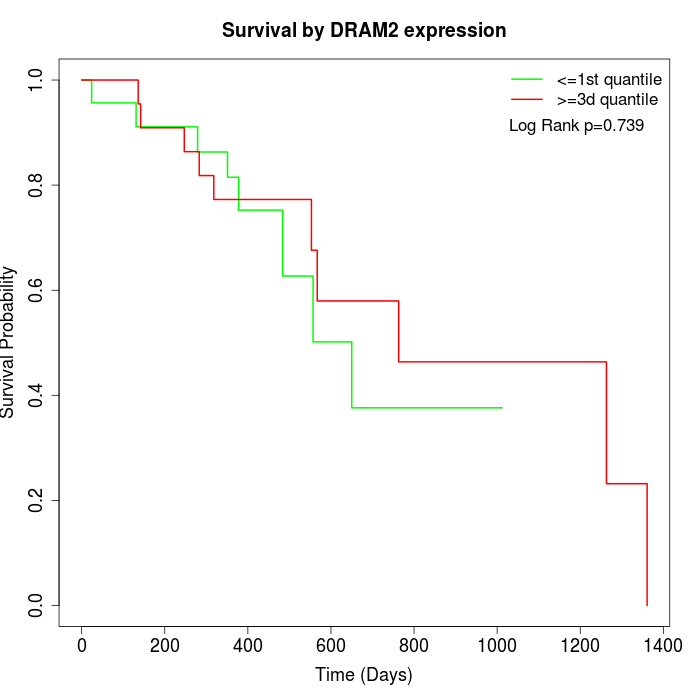
Survival by DRAM2 expression:
Note: Click image to view full size file.
Copy number change of DRAM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DRAM2 | 128338 | 0 | 11 | 19 | |
| GSE20123 | DRAM2 | 128338 | 0 | 11 | 19 | |
| GSE43470 | DRAM2 | 128338 | 0 | 8 | 35 | |
| GSE46452 | DRAM2 | 128338 | 2 | 1 | 56 | |
| GSE47630 | DRAM2 | 128338 | 9 | 5 | 26 | |
| GSE54993 | DRAM2 | 128338 | 0 | 1 | 69 | |
| GSE54994 | DRAM2 | 128338 | 7 | 4 | 42 | |
| GSE60625 | DRAM2 | 128338 | 0 | 0 | 11 | |
| GSE74703 | DRAM2 | 128338 | 0 | 7 | 29 | |
| GSE74704 | DRAM2 | 128338 | 0 | 7 | 13 | |
| TCGA | DRAM2 | 128338 | 7 | 27 | 62 |
Total number of gains: 25; Total number of losses: 82; Total Number of normals: 381.
Somatic mutations of DRAM2:
Generating mutation plots.
Highly correlated genes for DRAM2:
Showing top 20/245 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DRAM2 | NAGLU | 0.781011 | 3 | 0 | 3 |
| DRAM2 | OSTC | 0.76042 | 3 | 0 | 3 |
| DRAM2 | WDR77 | 0.758871 | 3 | 0 | 3 |
| DRAM2 | RAB18 | 0.753279 | 4 | 0 | 4 |
| DRAM2 | APPL2 | 0.752993 | 3 | 0 | 3 |
| DRAM2 | DCAF10 | 0.746675 | 3 | 0 | 3 |
| DRAM2 | PPWD1 | 0.743406 | 3 | 0 | 3 |
| DRAM2 | ATP6V1E1 | 0.742907 | 3 | 0 | 3 |
| DRAM2 | HDDC3 | 0.739677 | 3 | 0 | 3 |
| DRAM2 | C3orf38 | 0.739274 | 3 | 0 | 3 |
| DRAM2 | CYFIP1 | 0.738809 | 3 | 0 | 3 |
| DRAM2 | CCDC6 | 0.738482 | 3 | 0 | 3 |
| DRAM2 | SETX | 0.736495 | 3 | 0 | 3 |
| DRAM2 | SAMM50 | 0.725288 | 3 | 0 | 3 |
| DRAM2 | ZNF318 | 0.724655 | 3 | 0 | 3 |
| DRAM2 | ZBTB1 | 0.72008 | 3 | 0 | 3 |
| DRAM2 | BLOC1S2 | 0.719299 | 3 | 0 | 3 |
| DRAM2 | FBXO28 | 0.715179 | 3 | 0 | 3 |
| DRAM2 | KCNJ2 | 0.71383 | 3 | 0 | 3 |
| DRAM2 | IFT46 | 0.712781 | 3 | 0 | 3 |
For details and further investigation, click here