| Full name: alpha-N-acetylgalactosaminidase | Alias Symbol: D22S674 | ||
| Type: protein-coding gene | Cytoband: 22q13.2 | ||
| Entrez ID: 4668 | HGNC ID: HGNC:7631 | Ensembl Gene: ENSG00000198951 | OMIM ID: 104170 |
| Drug and gene relationship at DGIdb | |||
Expression of NAGA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NAGA | 4668 | 1555041_a_at | -0.3538 | 0.4115 | |
| GSE20347 | NAGA | 4668 | 202943_s_at | -0.3497 | 0.0022 | |
| GSE23400 | NAGA | 4668 | 202944_at | 0.1556 | 0.0022 | |
| GSE26886 | NAGA | 4668 | 1555041_a_at | -0.6533 | 0.0007 | |
| GSE29001 | NAGA | 4668 | 202944_at | -0.0148 | 0.9403 | |
| GSE38129 | NAGA | 4668 | 202943_s_at | -0.2285 | 0.0266 | |
| GSE45670 | NAGA | 4668 | 1555041_a_at | -0.0023 | 0.9899 | |
| GSE53622 | NAGA | 4668 | 30500 | 0.0864 | 0.2278 | |
| GSE53624 | NAGA | 4668 | 30500 | 0.0389 | 0.5569 | |
| GSE63941 | NAGA | 4668 | 1555041_a_at | 0.1968 | 0.7256 | |
| GSE77861 | NAGA | 4668 | 202943_s_at | -0.2544 | 0.1687 | |
| GSE97050 | NAGA | 4668 | A_33_P3349444 | 0.3618 | 0.2926 | |
| SRP007169 | NAGA | 4668 | RNAseq | -0.1005 | 0.8266 | |
| SRP008496 | NAGA | 4668 | RNAseq | -0.1779 | 0.4850 | |
| SRP064894 | NAGA | 4668 | RNAseq | 0.5450 | 0.0000 | |
| SRP133303 | NAGA | 4668 | RNAseq | 0.2069 | 0.1506 | |
| SRP159526 | NAGA | 4668 | RNAseq | 0.3120 | 0.0908 | |
| SRP193095 | NAGA | 4668 | RNAseq | 0.0708 | 0.4833 | |
| SRP219564 | NAGA | 4668 | RNAseq | 0.4692 | 0.2219 | |
| TCGA | NAGA | 4668 | RNAseq | 0.0560 | 0.2560 |
Upregulated datasets: 0; Downregulated datasets: 0.
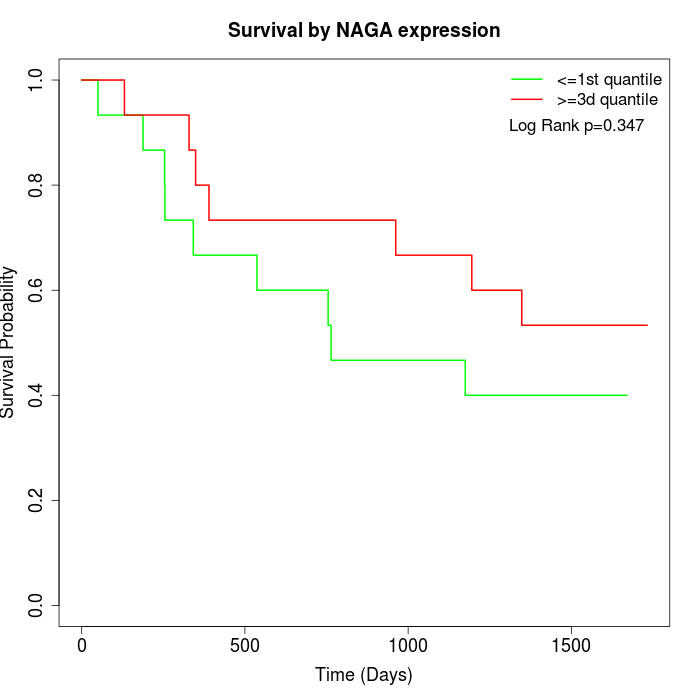
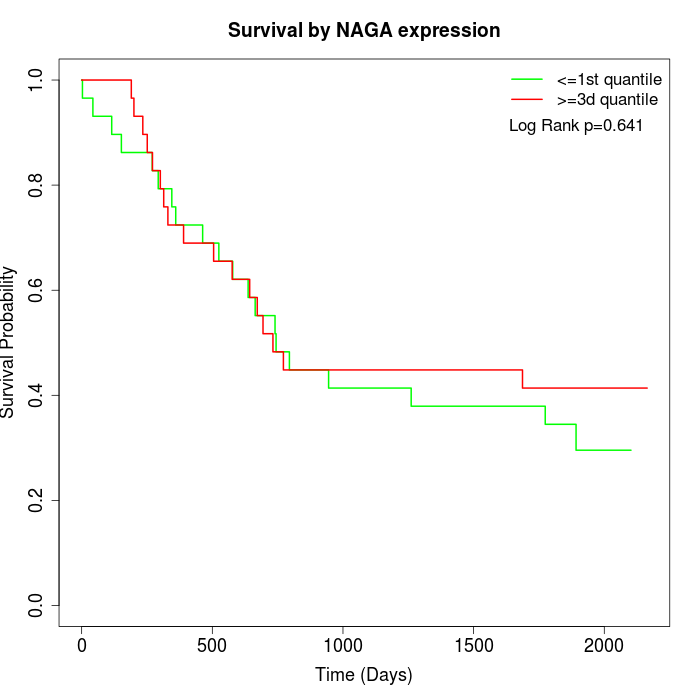
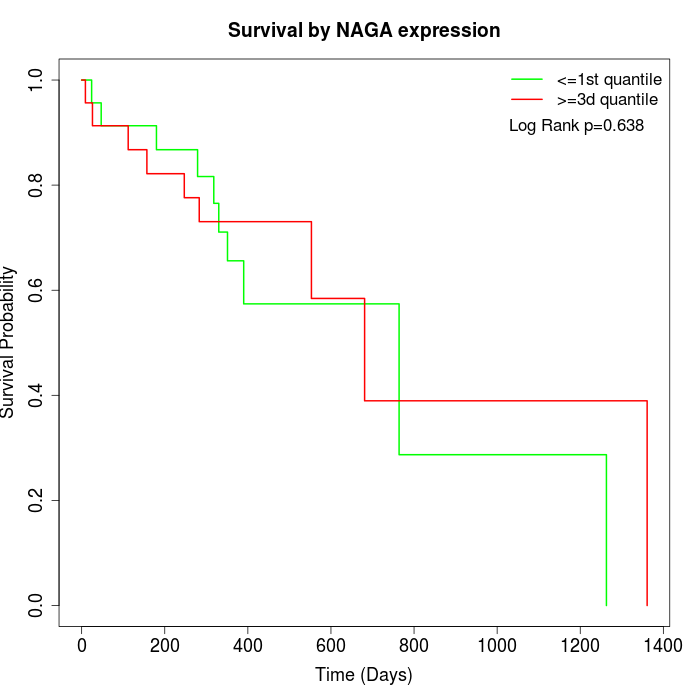
Survival by NAGA expression:
Note: Click image to view full size file.
Copy number change of NAGA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NAGA | 4668 | 6 | 5 | 19 | |
| GSE20123 | NAGA | 4668 | 6 | 4 | 20 | |
| GSE43470 | NAGA | 4668 | 5 | 6 | 32 | |
| GSE46452 | NAGA | 4668 | 31 | 2 | 26 | |
| GSE47630 | NAGA | 4668 | 9 | 4 | 27 | |
| GSE54993 | NAGA | 4668 | 3 | 6 | 61 | |
| GSE54994 | NAGA | 4668 | 11 | 8 | 34 | |
| GSE60625 | NAGA | 4668 | 5 | 0 | 6 | |
| GSE74703 | NAGA | 4668 | 5 | 4 | 27 | |
| GSE74704 | NAGA | 4668 | 3 | 2 | 15 | |
| TCGA | NAGA | 4668 | 27 | 14 | 55 |
Total number of gains: 111; Total number of losses: 55; Total Number of normals: 322.
Somatic mutations of NAGA:
Generating mutation plots.
Highly correlated genes for NAGA:
Showing top 20/421 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NAGA | PSMD8 | 0.798649 | 3 | 0 | 3 |
| NAGA | KIAA0355 | 0.797889 | 3 | 0 | 3 |
| NAGA | KCNJ2 | 0.793674 | 3 | 0 | 3 |
| NAGA | MAT2B | 0.744424 | 3 | 0 | 3 |
| NAGA | ZFP36L2 | 0.736638 | 3 | 0 | 3 |
| NAGA | SERTAD1 | 0.732191 | 3 | 0 | 3 |
| NAGA | SPRYD4 | 0.727516 | 3 | 0 | 3 |
| NAGA | PFDN1 | 0.719496 | 3 | 0 | 3 |
| NAGA | SLC35A4 | 0.718355 | 4 | 0 | 4 |
| NAGA | MAP3K1 | 0.712521 | 4 | 0 | 4 |
| NAGA | LMO2 | 0.712191 | 4 | 0 | 3 |
| NAGA | PTGR1 | 0.710159 | 4 | 0 | 3 |
| NAGA | FAM122A | 0.705937 | 4 | 0 | 4 |
| NAGA | P2RX7 | 0.705828 | 3 | 0 | 3 |
| NAGA | ABI1 | 0.701976 | 3 | 0 | 3 |
| NAGA | ABHD14B | 0.698477 | 3 | 0 | 3 |
| NAGA | TAB1 | 0.693398 | 3 | 0 | 3 |
| NAGA | BNIP1 | 0.69335 | 3 | 0 | 3 |
| NAGA | SSNA1 | 0.689636 | 4 | 0 | 3 |
| NAGA | OGFRL1 | 0.689524 | 3 | 0 | 3 |
For details and further investigation, click here