| Full name: long intergenic non-protein coding RNA 347 | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 13q22.1 | ||
| Entrez ID: 338864 | HGNC ID: HGNC:27890 | Ensembl Gene: ENSG00000236678 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LINC00347:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LINC00347 | 338864 | 237980_at | -0.0299 | 0.9064 | |
| GSE26886 | LINC00347 | 338864 | 237980_at | -0.2520 | 0.0361 | |
| GSE45670 | LINC00347 | 338864 | 237980_at | 0.0726 | 0.3504 | |
| GSE53622 | LINC00347 | 338864 | 10038 | 0.4000 | 0.0010 | |
| GSE53624 | LINC00347 | 338864 | 89270 | 0.1612 | 0.1593 | |
| GSE63941 | LINC00347 | 338864 | 237980_at | 0.1886 | 0.1728 | |
| GSE77861 | LINC00347 | 338864 | 237980_at | -0.1001 | 0.2912 |
Upregulated datasets: 0; Downregulated datasets: 0.
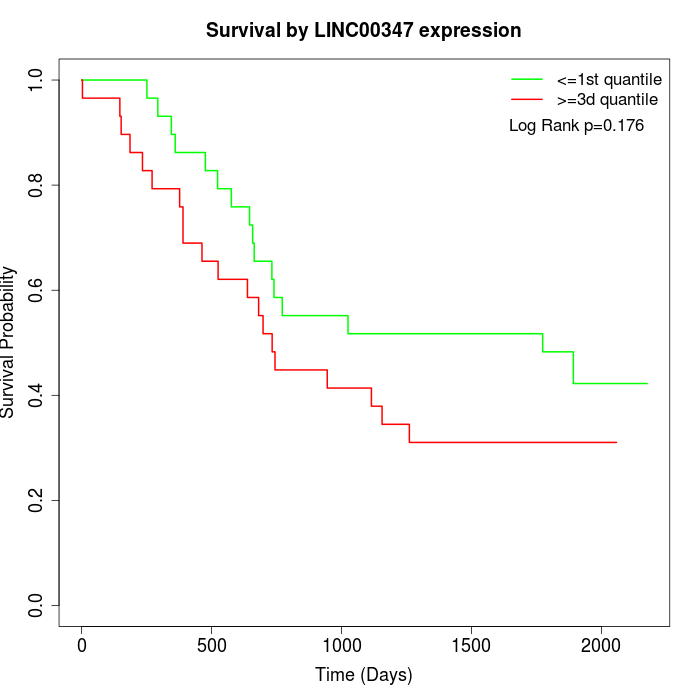
Survival by LINC00347 expression:
Note: Click image to view full size file.
Copy number change of LINC00347:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LINC00347 | 338864 | 3 | 10 | 17 | |
| GSE20123 | LINC00347 | 338864 | 3 | 9 | 18 | |
| GSE43470 | LINC00347 | 338864 | 4 | 10 | 29 | |
| GSE46452 | LINC00347 | 338864 | 0 | 32 | 27 | |
| GSE47630 | LINC00347 | 338864 | 3 | 27 | 10 | |
| GSE54993 | LINC00347 | 338864 | 11 | 3 | 56 | |
| GSE54994 | LINC00347 | 338864 | 4 | 13 | 36 | |
| GSE60625 | LINC00347 | 338864 | 0 | 3 | 8 | |
| GSE74703 | LINC00347 | 338864 | 3 | 8 | 25 | |
| GSE74704 | LINC00347 | 338864 | 1 | 8 | 11 | |
| TCGA | LINC00347 | 338864 | 12 | 34 | 50 |
Total number of gains: 44; Total number of losses: 157; Total Number of normals: 287.
Somatic mutations of LINC00347:
Generating mutation plots.
Highly correlated genes for LINC00347:
Showing top 20/104 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LINC00347 | KCNQ1DN | 0.705443 | 3 | 0 | 3 |
| LINC00347 | PRB1 | 0.700985 | 4 | 0 | 4 |
| LINC00347 | GPR45 | 0.67371 | 3 | 0 | 3 |
| LINC00347 | KCNK16 | 0.669099 | 3 | 0 | 3 |
| LINC00347 | GAPDHS | 0.66023 | 3 | 0 | 3 |
| LINC00347 | C2orf16 | 0.659524 | 4 | 0 | 3 |
| LINC00347 | AVPR1B | 0.659339 | 3 | 0 | 3 |
| LINC00347 | SLC22A14 | 0.652201 | 5 | 0 | 4 |
| LINC00347 | HIF3A | 0.651101 | 3 | 0 | 3 |
| LINC00347 | ARX | 0.639479 | 3 | 0 | 3 |
| LINC00347 | SHISA9 | 0.633279 | 4 | 0 | 3 |
| LINC00347 | LINC01119 | 0.629316 | 3 | 0 | 3 |
| LINC00347 | PRSS33 | 0.628453 | 3 | 0 | 3 |
| LINC00347 | TMPRSS15 | 0.6277 | 3 | 0 | 3 |
| LINC00347 | SLC8A3 | 0.626583 | 3 | 0 | 3 |
| LINC00347 | CA1 | 0.62493 | 4 | 0 | 3 |
| LINC00347 | TEX28 | 0.623536 | 5 | 0 | 4 |
| LINC00347 | TXNDC2 | 0.619197 | 5 | 0 | 3 |
| LINC00347 | ZFHX2 | 0.609072 | 3 | 0 | 3 |
| LINC00347 | DOK3 | 0.606616 | 4 | 0 | 3 |
For details and further investigation, click here