| Full name: LIM zinc finger domain containing 2 | Alias Symbol: PINCH2|PINCH-2 | ||
| Type: protein-coding gene | Cytoband: 2q14.3 | ||
| Entrez ID: 55679 | HGNC ID: HGNC:16084 | Ensembl Gene: ENSG00000072163 | OMIM ID: 607908 |
| Drug and gene relationship at DGIdb | |||
Expression of LIMS2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LIMS2 | 55679 | 220765_s_at | -0.8388 | 0.3855 | |
| GSE20347 | LIMS2 | 55679 | 220765_s_at | -0.0551 | 0.7831 | |
| GSE23400 | LIMS2 | 55679 | 220765_s_at | -0.5192 | 0.0012 | |
| GSE26886 | LIMS2 | 55679 | 220765_s_at | 0.2621 | 0.1221 | |
| GSE29001 | LIMS2 | 55679 | 220765_s_at | -0.1575 | 0.5501 | |
| GSE38129 | LIMS2 | 55679 | 220765_s_at | -0.7472 | 0.0178 | |
| GSE45670 | LIMS2 | 55679 | 220765_s_at | -0.6172 | 0.0000 | |
| GSE53622 | LIMS2 | 55679 | 57831 | -1.5575 | 0.0000 | |
| GSE53624 | LIMS2 | 55679 | 161833 | -1.2037 | 0.0000 | |
| GSE63941 | LIMS2 | 55679 | 220765_s_at | -0.9934 | 0.0038 | |
| GSE77861 | LIMS2 | 55679 | 220765_s_at | -0.0669 | 0.5212 | |
| GSE97050 | LIMS2 | 55679 | A_33_P3268181 | -1.3322 | 0.1399 | |
| SRP007169 | LIMS2 | 55679 | RNAseq | 1.6071 | 0.0101 | |
| SRP008496 | LIMS2 | 55679 | RNAseq | 1.9602 | 0.0009 | |
| SRP064894 | LIMS2 | 55679 | RNAseq | -0.8209 | 0.0180 | |
| SRP133303 | LIMS2 | 55679 | RNAseq | -1.6554 | 0.0000 | |
| SRP159526 | LIMS2 | 55679 | RNAseq | -1.1297 | 0.0462 | |
| SRP219564 | LIMS2 | 55679 | RNAseq | -0.9454 | 0.3586 | |
| TCGA | LIMS2 | 55679 | RNAseq | -0.6081 | 0.0000 |
Upregulated datasets: 2; Downregulated datasets: 4.
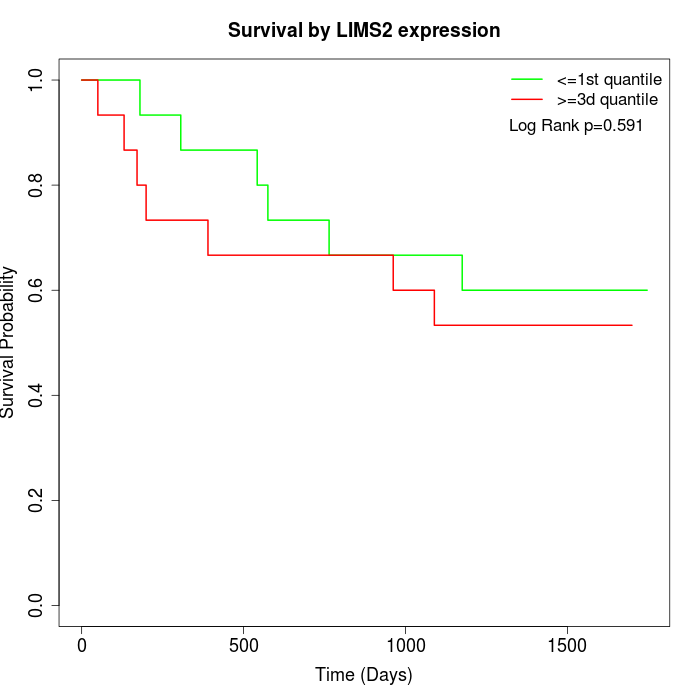
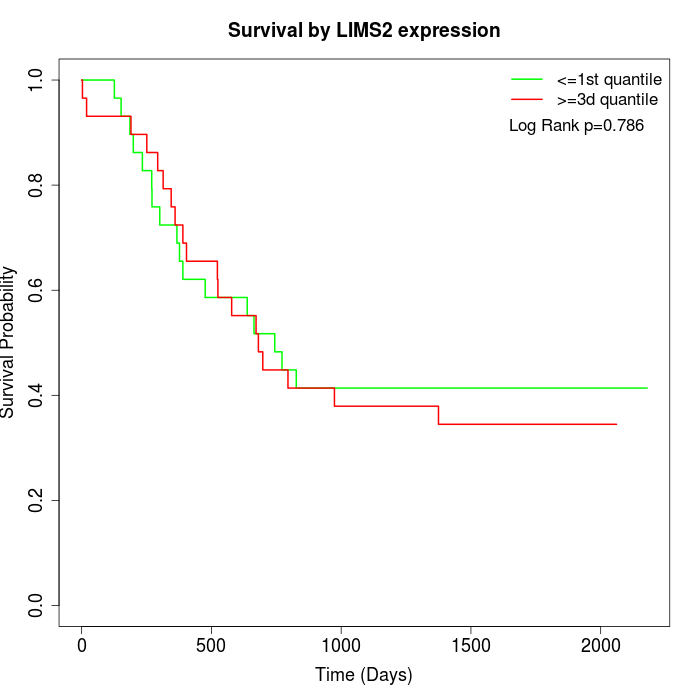
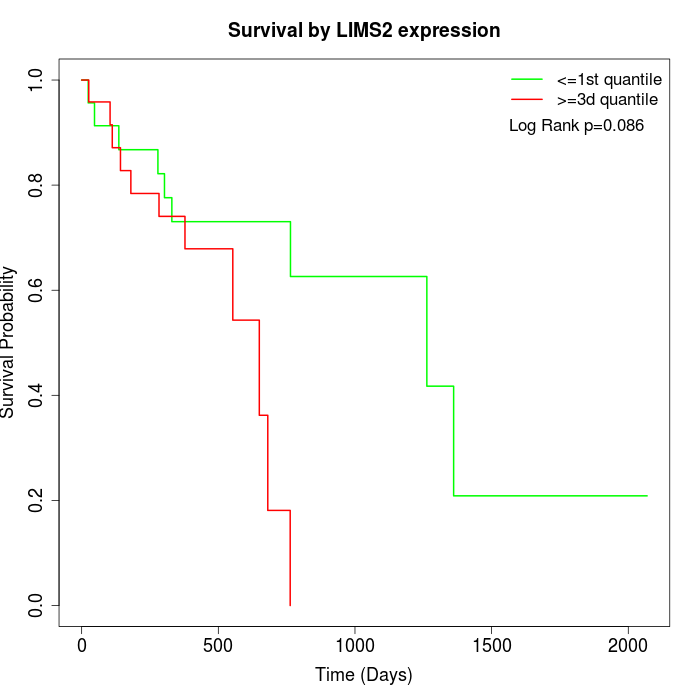
Survival by LIMS2 expression:
Note: Click image to view full size file.
Copy number change of LIMS2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LIMS2 | 55679 | 6 | 3 | 21 | |
| GSE20123 | LIMS2 | 55679 | 6 | 3 | 21 | |
| GSE43470 | LIMS2 | 55679 | 2 | 1 | 40 | |
| GSE46452 | LIMS2 | 55679 | 1 | 3 | 55 | |
| GSE47630 | LIMS2 | 55679 | 6 | 0 | 34 | |
| GSE54993 | LIMS2 | 55679 | 0 | 6 | 64 | |
| GSE54994 | LIMS2 | 55679 | 11 | 0 | 42 | |
| GSE60625 | LIMS2 | 55679 | 0 | 3 | 8 | |
| GSE74703 | LIMS2 | 55679 | 2 | 1 | 33 | |
| GSE74704 | LIMS2 | 55679 | 3 | 1 | 16 | |
| TCGA | LIMS2 | 55679 | 22 | 8 | 66 |
Total number of gains: 59; Total number of losses: 29; Total Number of normals: 400.
Somatic mutations of LIMS2:
Generating mutation plots.
Highly correlated genes for LIMS2:
Showing top 20/1117 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LIMS2 | UBXN10 | 0.881612 | 3 | 0 | 3 |
| LIMS2 | SORCS1 | 0.848693 | 4 | 0 | 4 |
| LIMS2 | MRVI1 | 0.837305 | 7 | 0 | 6 |
| LIMS2 | MYOCD | 0.822575 | 7 | 0 | 7 |
| LIMS2 | CLEC3B | 0.81148 | 3 | 0 | 3 |
| LIMS2 | BHMT2 | 0.808163 | 11 | 0 | 11 |
| LIMS2 | TLN1 | 0.807103 | 6 | 0 | 6 |
| LIMS2 | CCDC69 | 0.805324 | 9 | 0 | 9 |
| LIMS2 | ITGA7 | 0.801873 | 10 | 0 | 9 |
| LIMS2 | ARHGAP6 | 0.788842 | 10 | 0 | 10 |
| LIMS2 | PPP1R12B | 0.786743 | 10 | 0 | 9 |
| LIMS2 | VMAC | 0.786611 | 3 | 0 | 3 |
| LIMS2 | ASB5 | 0.785827 | 6 | 0 | 6 |
| LIMS2 | FLNC | 0.785128 | 12 | 0 | 11 |
| LIMS2 | KCNMA1 | 0.782896 | 10 | 0 | 9 |
| LIMS2 | MYOC | 0.782871 | 9 | 0 | 8 |
| LIMS2 | STARD9 | 0.779783 | 6 | 0 | 6 |
| LIMS2 | MECP2 | 0.779642 | 5 | 0 | 5 |
| LIMS2 | LMOD1 | 0.778337 | 12 | 0 | 11 |
| LIMS2 | SGCA | 0.776467 | 12 | 0 | 12 |
For details and further investigation, click here