| Full name: lipase F, gastric type | Alias Symbol: HGL|HLAL | ||
| Type: protein-coding gene | Cytoband: 10q23.31 | ||
| Entrez ID: 8513 | HGNC ID: HGNC:6622 | Ensembl Gene: ENSG00000182333 | OMIM ID: 601980 |
| Drug and gene relationship at DGIdb | |||
Expression of LIPF:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LIPF | 8513 | 206334_at | -0.1136 | 0.5807 | |
| GSE20347 | LIPF | 8513 | 206334_at | -0.3477 | 0.4222 | |
| GSE23400 | LIPF | 8513 | 206334_at | -0.4139 | 0.1667 | |
| GSE26886 | LIPF | 8513 | 206334_at | -0.8661 | 0.1064 | |
| GSE29001 | LIPF | 8513 | 206334_at | -0.0185 | 0.8919 | |
| GSE38129 | LIPF | 8513 | 206334_at | -1.2702 | 0.0677 | |
| GSE45670 | LIPF | 8513 | 206334_at | 0.1752 | 0.0706 | |
| GSE53622 | LIPF | 8513 | 116570 | -0.2156 | 0.6720 | |
| GSE53624 | LIPF | 8513 | 116570 | -0.6945 | 0.1381 | |
| GSE63941 | LIPF | 8513 | 206334_at | -0.0050 | 0.9712 | |
| GSE77861 | LIPF | 8513 | 206334_at | -0.0127 | 0.8969 | |
| GSE97050 | LIPF | 8513 | A_23_P47019 | -0.1652 | 0.4222 | |
| TCGA | LIPF | 8513 | RNAseq | -5.7729 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 1.
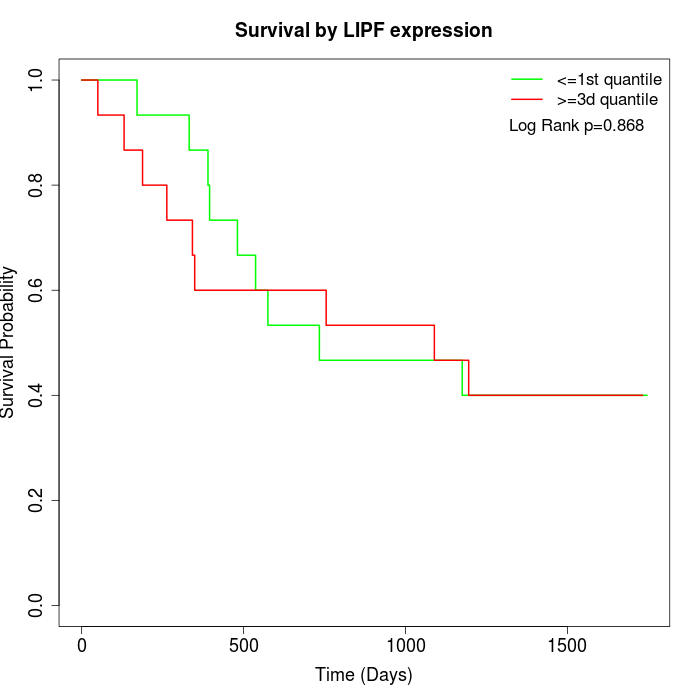
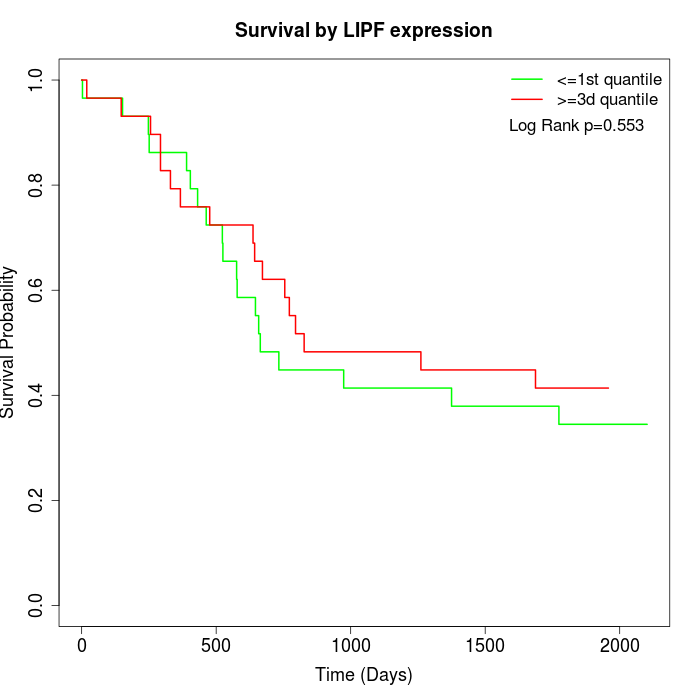
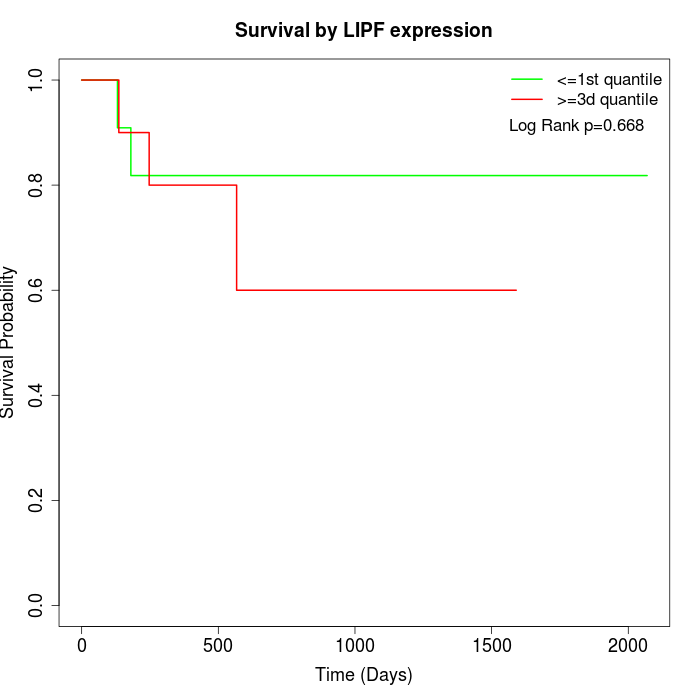
Survival by LIPF expression:
Note: Click image to view full size file.
Copy number change of LIPF:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LIPF | 8513 | 2 | 7 | 21 | |
| GSE20123 | LIPF | 8513 | 2 | 6 | 22 | |
| GSE43470 | LIPF | 8513 | 0 | 8 | 35 | |
| GSE46452 | LIPF | 8513 | 0 | 11 | 48 | |
| GSE47630 | LIPF | 8513 | 2 | 14 | 24 | |
| GSE54993 | LIPF | 8513 | 7 | 0 | 63 | |
| GSE54994 | LIPF | 8513 | 1 | 11 | 41 | |
| GSE60625 | LIPF | 8513 | 0 | 0 | 11 | |
| GSE74703 | LIPF | 8513 | 0 | 6 | 30 | |
| GSE74704 | LIPF | 8513 | 1 | 4 | 15 | |
| TCGA | LIPF | 8513 | 7 | 27 | 62 |
Total number of gains: 22; Total number of losses: 94; Total Number of normals: 372.
Somatic mutations of LIPF:
Generating mutation plots.
Highly correlated genes for LIPF:
Showing top 20/323 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LIPF | RARA | 0.866724 | 3 | 0 | 3 |
| LIPF | ADAM11 | 0.826025 | 4 | 0 | 4 |
| LIPF | SLC13A2 | 0.813388 | 3 | 0 | 3 |
| LIPF | ESR2 | 0.799694 | 3 | 0 | 3 |
| LIPF | CLDN18 | 0.79343 | 8 | 0 | 7 |
| LIPF | GKN1 | 0.790659 | 7 | 0 | 7 |
| LIPF | CA5B | 0.783157 | 3 | 0 | 3 |
| LIPF | PHLDB1 | 0.782722 | 4 | 0 | 4 |
| LIPF | MAPRE3 | 0.77729 | 3 | 0 | 3 |
| LIPF | BAGE | 0.77619 | 3 | 0 | 3 |
| LIPF | ENTPD2 | 0.7684 | 3 | 0 | 3 |
| LIPF | RNF207 | 0.767421 | 3 | 0 | 3 |
| LIPF | IRS4 | 0.764994 | 3 | 0 | 3 |
| LIPF | NLGN4Y | 0.763359 | 3 | 0 | 3 |
| LIPF | SPINK1 | 0.761724 | 6 | 0 | 6 |
| LIPF | MAPK11 | 0.760849 | 3 | 0 | 3 |
| LIPF | RXRG | 0.75658 | 3 | 0 | 3 |
| LIPF | NELL1 | 0.755921 | 3 | 0 | 3 |
| LIPF | PRPH2 | 0.750218 | 3 | 0 | 3 |
| LIPF | GRIA4 | 0.749972 | 3 | 0 | 3 |
For details and further investigation, click here