| Full name: potassium sodium-activated channel subfamily T member 1 | Alias Symbol: KCa4.1|KIAA1422|SLACK|Slo2.2 | ||
| Type: protein-coding gene | Cytoband: 9q34.3 | ||
| Entrez ID: 57582 | HGNC ID: HGNC:18865 | Ensembl Gene: ENSG00000107147 | OMIM ID: 608167 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNT1 | 57582 | 1569461_at | -0.0840 | 0.7523 | |
| GSE26886 | KCNT1 | 57582 | 1569461_at | 0.0147 | 0.9431 | |
| GSE45670 | KCNT1 | 57582 | 1569461_at | -0.0293 | 0.7765 | |
| GSE53622 | KCNT1 | 57582 | 49849 | -0.2928 | 0.0743 | |
| GSE53624 | KCNT1 | 57582 | 49849 | -0.9968 | 0.0000 | |
| GSE63941 | KCNT1 | 57582 | 1569461_at | -0.0477 | 0.7621 | |
| GSE77861 | KCNT1 | 57582 | 1569461_at | -0.1982 | 0.0966 | |
| GSE97050 | KCNT1 | 57582 | A_33_P3281613 | 0.0363 | 0.8840 | |
| SRP133303 | KCNT1 | 57582 | RNAseq | -0.5818 | 0.0125 | |
| SRP159526 | KCNT1 | 57582 | RNAseq | -0.0209 | 0.9723 | |
| SRP219564 | KCNT1 | 57582 | RNAseq | -0.4798 | 0.1918 | |
| TCGA | KCNT1 | 57582 | RNAseq | -1.5710 | 0.0045 |
Upregulated datasets: 0; Downregulated datasets: 1.
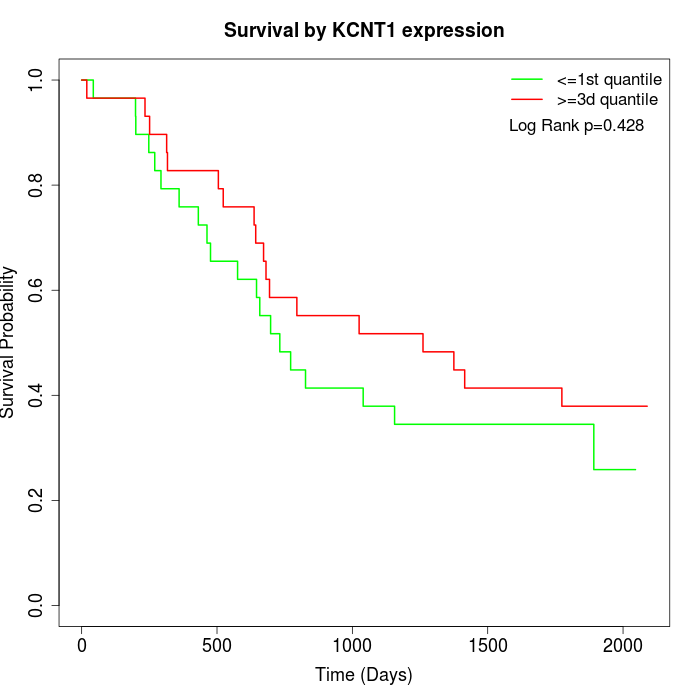
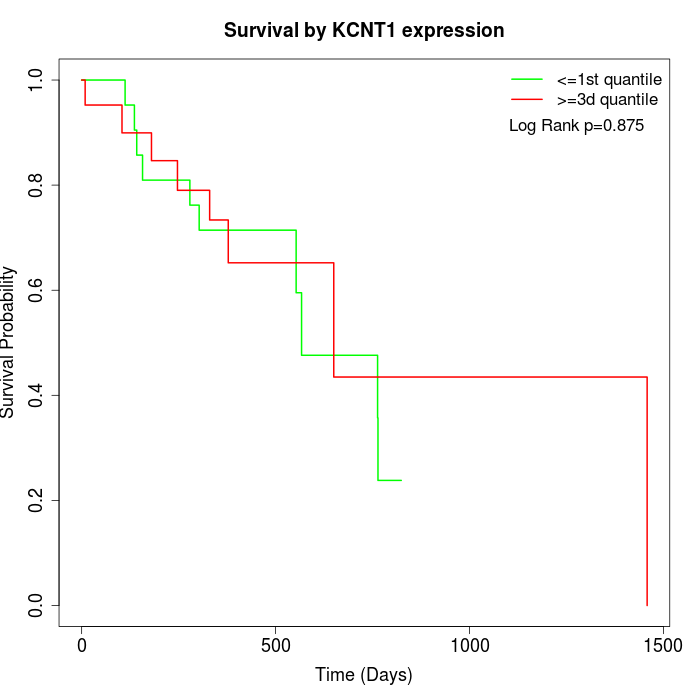
Survival by KCNT1 expression:
Note: Click image to view full size file.
Copy number change of KCNT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNT1 | 57582 | 5 | 7 | 18 | |
| GSE20123 | KCNT1 | 57582 | 5 | 7 | 18 | |
| GSE43470 | KCNT1 | 57582 | 3 | 7 | 33 | |
| GSE46452 | KCNT1 | 57582 | 6 | 13 | 40 | |
| GSE47630 | KCNT1 | 57582 | 6 | 15 | 19 | |
| GSE54993 | KCNT1 | 57582 | 3 | 3 | 64 | |
| GSE54994 | KCNT1 | 57582 | 12 | 8 | 33 | |
| GSE60625 | KCNT1 | 57582 | 0 | 0 | 11 | |
| GSE74703 | KCNT1 | 57582 | 3 | 5 | 28 | |
| GSE74704 | KCNT1 | 57582 | 3 | 5 | 12 | |
| TCGA | KCNT1 | 57582 | 28 | 25 | 43 |
Total number of gains: 74; Total number of losses: 95; Total Number of normals: 319.
Somatic mutations of KCNT1:
Generating mutation plots.
Highly correlated genes for KCNT1:
Showing top 20/315 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNT1 | IGFN1 | 0.839089 | 3 | 0 | 3 |
| KCNT1 | HOXD9 | 0.812482 | 3 | 0 | 3 |
| KCNT1 | CD300LG | 0.801884 | 3 | 0 | 3 |
| KCNT1 | TMEM37 | 0.795353 | 3 | 0 | 3 |
| KCNT1 | FAM171A2 | 0.784193 | 4 | 0 | 4 |
| KCNT1 | TMEM52 | 0.779171 | 3 | 0 | 3 |
| KCNT1 | SEC13 | 0.769789 | 3 | 0 | 3 |
| KCNT1 | SHANK1 | 0.764839 | 4 | 0 | 4 |
| KCNT1 | IGSF21 | 0.755201 | 3 | 0 | 3 |
| KCNT1 | ZMYND10 | 0.752058 | 4 | 0 | 4 |
| KCNT1 | ENTPD5 | 0.74913 | 3 | 0 | 3 |
| KCNT1 | GALR2 | 0.747912 | 3 | 0 | 3 |
| KCNT1 | CTSE | 0.745341 | 3 | 0 | 3 |
| KCNT1 | MYH14 | 0.743792 | 3 | 0 | 3 |
| KCNT1 | CARD6 | 0.740354 | 3 | 0 | 3 |
| KCNT1 | FAM71A | 0.729727 | 4 | 0 | 4 |
| KCNT1 | CEBPE | 0.729597 | 3 | 0 | 3 |
| KCNT1 | TNR | 0.729319 | 4 | 0 | 4 |
| KCNT1 | RIPK4 | 0.725176 | 3 | 0 | 3 |
| KCNT1 | MAG | 0.723191 | 3 | 0 | 3 |
For details and further investigation, click here