| Full name: potassium channel tetramerization domain containing 17 | Alias Symbol: FLJ12242 | ||
| Type: protein-coding gene | Cytoband: 22q12.3 | ||
| Entrez ID: 79734 | HGNC ID: HGNC:25705 | Ensembl Gene: ENSG00000100379 | OMIM ID: 616386 |
| Drug and gene relationship at DGIdb | |||
Expression of KCTD17:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCTD17 | 79734 | 205561_at | 0.1033 | 0.7318 | |
| GSE20347 | KCTD17 | 79734 | 205561_at | 0.1912 | 0.0077 | |
| GSE23400 | KCTD17 | 79734 | 205561_at | -0.0610 | 0.1453 | |
| GSE26886 | KCTD17 | 79734 | 205561_at | 0.3901 | 0.0019 | |
| GSE29001 | KCTD17 | 79734 | 205561_at | 0.0843 | 0.7288 | |
| GSE38129 | KCTD17 | 79734 | 205561_at | 0.1090 | 0.1789 | |
| GSE45670 | KCTD17 | 79734 | 205561_at | 0.0077 | 0.9429 | |
| GSE53622 | KCTD17 | 79734 | 55695 | 0.2311 | 0.0361 | |
| GSE53624 | KCTD17 | 79734 | 55695 | 0.4631 | 0.0000 | |
| GSE63941 | KCTD17 | 79734 | 205561_at | -0.4113 | 0.0418 | |
| GSE77861 | KCTD17 | 79734 | 205561_at | -0.0302 | 0.8559 | |
| GSE97050 | KCTD17 | 79734 | A_24_P108779 | -0.2336 | 0.4907 | |
| SRP007169 | KCTD17 | 79734 | RNAseq | -0.2985 | 0.6212 | |
| SRP008496 | KCTD17 | 79734 | RNAseq | -0.1419 | 0.7266 | |
| SRP064894 | KCTD17 | 79734 | RNAseq | 0.6261 | 0.0709 | |
| SRP133303 | KCTD17 | 79734 | RNAseq | 0.5622 | 0.0379 | |
| SRP159526 | KCTD17 | 79734 | RNAseq | 0.8737 | 0.0020 | |
| SRP193095 | KCTD17 | 79734 | RNAseq | 0.9142 | 0.0000 | |
| SRP219564 | KCTD17 | 79734 | RNAseq | 0.3321 | 0.5867 | |
| TCGA | KCTD17 | 79734 | RNAseq | 0.2798 | 0.0088 |
Upregulated datasets: 0; Downregulated datasets: 0.
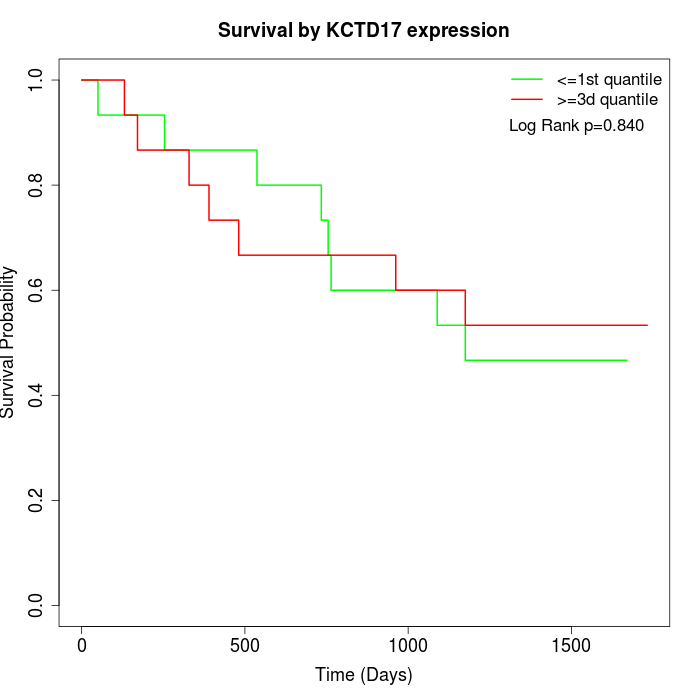
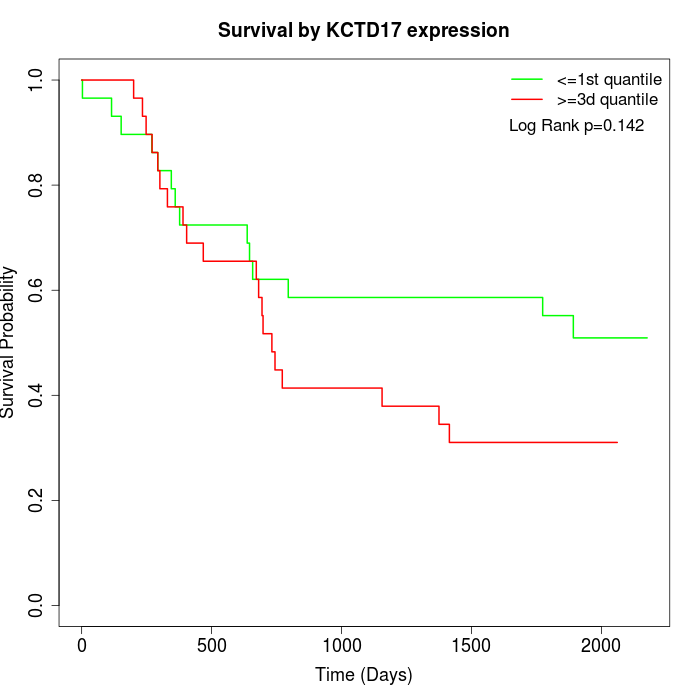
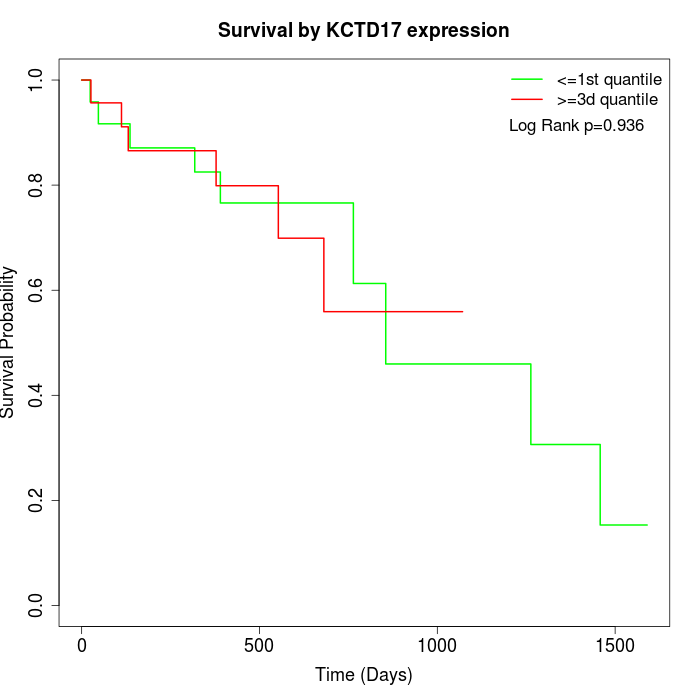
Survival by KCTD17 expression:
Note: Click image to view full size file.
Copy number change of KCTD17:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCTD17 | 79734 | 6 | 6 | 18 | |
| GSE20123 | KCTD17 | 79734 | 6 | 5 | 19 | |
| GSE43470 | KCTD17 | 79734 | 4 | 6 | 33 | |
| GSE46452 | KCTD17 | 79734 | 31 | 1 | 27 | |
| GSE47630 | KCTD17 | 79734 | 8 | 5 | 27 | |
| GSE54993 | KCTD17 | 79734 | 3 | 6 | 61 | |
| GSE54994 | KCTD17 | 79734 | 10 | 8 | 35 | |
| GSE60625 | KCTD17 | 79734 | 5 | 0 | 6 | |
| GSE74703 | KCTD17 | 79734 | 4 | 4 | 28 | |
| GSE74704 | KCTD17 | 79734 | 3 | 2 | 15 | |
| TCGA | KCTD17 | 79734 | 27 | 16 | 53 |
Total number of gains: 107; Total number of losses: 59; Total Number of normals: 322.
Somatic mutations of KCTD17:
Generating mutation plots.
Highly correlated genes for KCTD17:
Showing top 20/444 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCTD17 | STX1A | 0.724161 | 3 | 0 | 3 |
| KCTD17 | PNCK | 0.6926 | 3 | 0 | 3 |
| KCTD17 | CNPY4 | 0.675856 | 6 | 0 | 6 |
| KCTD17 | C8orf44 | 0.67574 | 3 | 0 | 3 |
| KCTD17 | TNK2 | 0.667155 | 5 | 0 | 5 |
| KCTD17 | DNM2 | 0.665634 | 3 | 0 | 3 |
| KCTD17 | ZNF536 | 0.659025 | 3 | 0 | 3 |
| KCTD17 | SIX1 | 0.65849 | 3 | 0 | 3 |
| KCTD17 | PI16 | 0.647879 | 4 | 0 | 3 |
| KCTD17 | AZIN2 | 0.643276 | 5 | 0 | 5 |
| KCTD17 | GRASP | 0.642026 | 5 | 0 | 3 |
| KCTD17 | COX7A1 | 0.638499 | 3 | 0 | 3 |
| KCTD17 | PRRX2 | 0.636946 | 5 | 0 | 4 |
| KCTD17 | ZNF358 | 0.636465 | 7 | 0 | 6 |
| KCTD17 | SLC9A1 | 0.636206 | 4 | 0 | 3 |
| KCTD17 | TRIM47 | 0.63553 | 3 | 0 | 3 |
| KCTD17 | TSPYL2 | 0.634153 | 5 | 0 | 4 |
| KCTD17 | ALOX15 | 0.634138 | 3 | 0 | 3 |
| KCTD17 | LINC00311 | 0.630377 | 3 | 0 | 3 |
| KCTD17 | ROPN1L | 0.626669 | 3 | 0 | 3 |
For details and further investigation, click here