| Full name: canopy FGF signaling regulator 4 | Alias Symbol: MGC40499|PRAT4B | ||
| Type: protein-coding gene | Cytoband: 7q22.1 | ||
| Entrez ID: 245812 | HGNC ID: HGNC:28631 | Ensembl Gene: ENSG00000166997 | OMIM ID: 610047 |
| Drug and gene relationship at DGIdb | |||
Expression of CNPY4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CNPY4 | 245812 | 227313_at | 0.1065 | 0.8393 | |
| GSE20347 | CNPY4 | 245812 | 217700_at | 0.0419 | 0.6911 | |
| GSE23400 | CNPY4 | 245812 | 217700_at | -0.0468 | 0.2370 | |
| GSE26886 | CNPY4 | 245812 | 227313_at | 0.4255 | 0.0221 | |
| GSE29001 | CNPY4 | 245812 | 217700_at | -0.0585 | 0.6775 | |
| GSE38129 | CNPY4 | 245812 | 217700_at | -0.0764 | 0.4222 | |
| GSE45670 | CNPY4 | 245812 | 227313_at | 0.1394 | 0.4923 | |
| GSE53622 | CNPY4 | 245812 | 96535 | 0.4864 | 0.0000 | |
| GSE53624 | CNPY4 | 245812 | 96535 | 0.7032 | 0.0000 | |
| GSE63941 | CNPY4 | 245812 | 227313_at | -1.6118 | 0.0028 | |
| GSE77861 | CNPY4 | 245812 | 217700_at | -0.0838 | 0.7212 | |
| GSE97050 | CNPY4 | 245812 | A_23_P157038 | 0.0971 | 0.8698 | |
| SRP007169 | CNPY4 | 245812 | RNAseq | 0.5403 | 0.3947 | |
| SRP008496 | CNPY4 | 245812 | RNAseq | 1.2008 | 0.0251 | |
| SRP064894 | CNPY4 | 245812 | RNAseq | 1.0377 | 0.0000 | |
| SRP133303 | CNPY4 | 245812 | RNAseq | 0.4681 | 0.0689 | |
| SRP159526 | CNPY4 | 245812 | RNAseq | 1.1790 | 0.0001 | |
| SRP193095 | CNPY4 | 245812 | RNAseq | 0.9035 | 0.0000 | |
| SRP219564 | CNPY4 | 245812 | RNAseq | 0.6849 | 0.2111 | |
| TCGA | CNPY4 | 245812 | RNAseq | 0.2984 | 0.0042 |
Upregulated datasets: 3; Downregulated datasets: 1.
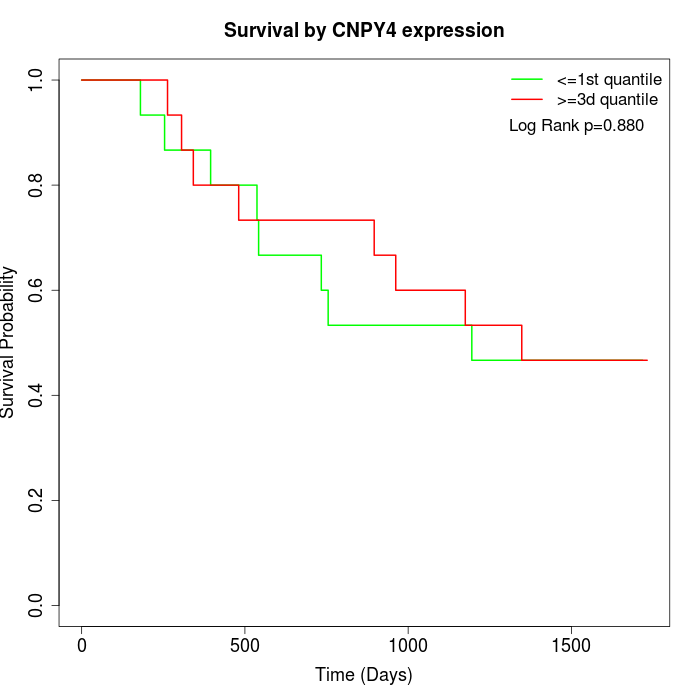
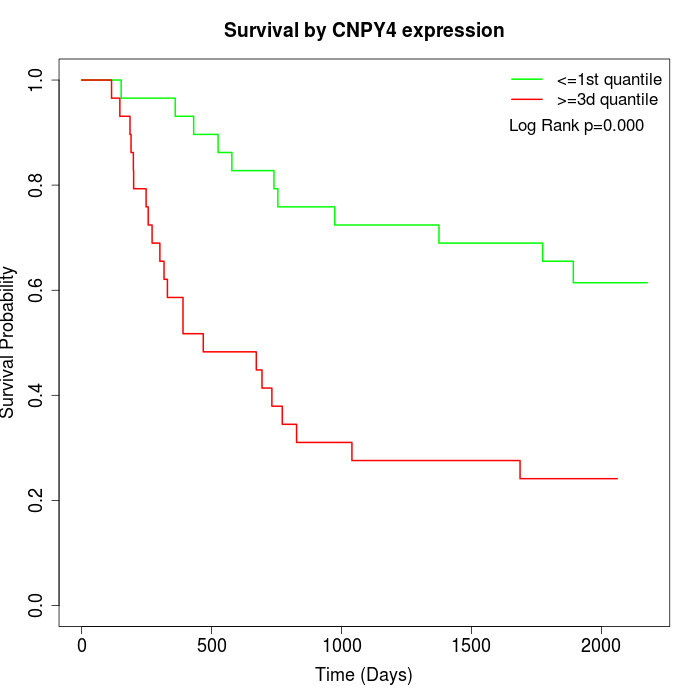
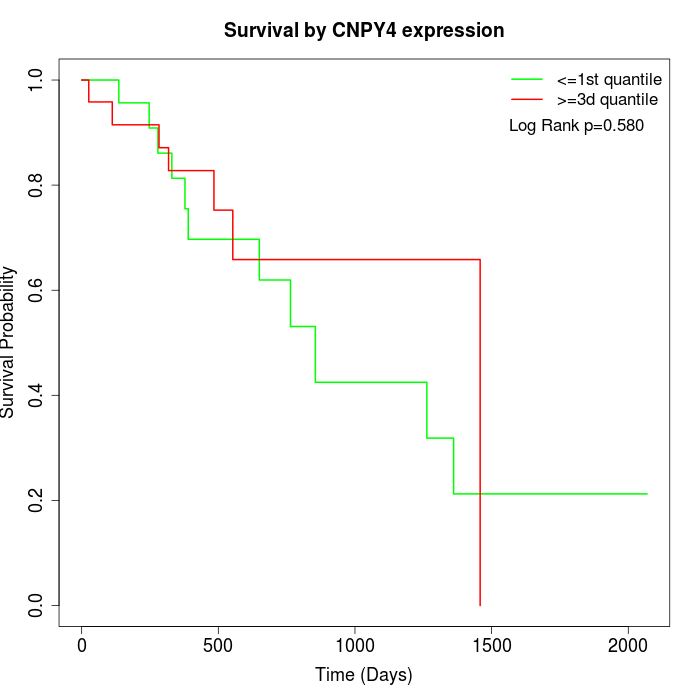
Survival by CNPY4 expression:
Note: Click image to view full size file.
Copy number change of CNPY4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CNPY4 | 245812 | 13 | 0 | 17 | |
| GSE20123 | CNPY4 | 245812 | 13 | 0 | 17 | |
| GSE43470 | CNPY4 | 245812 | 6 | 2 | 35 | |
| GSE46452 | CNPY4 | 245812 | 11 | 1 | 47 | |
| GSE47630 | CNPY4 | 245812 | 7 | 3 | 30 | |
| GSE54993 | CNPY4 | 245812 | 1 | 9 | 60 | |
| GSE54994 | CNPY4 | 245812 | 15 | 3 | 35 | |
| GSE60625 | CNPY4 | 245812 | 0 | 0 | 11 | |
| GSE74703 | CNPY4 | 245812 | 6 | 1 | 29 | |
| GSE74704 | CNPY4 | 245812 | 9 | 0 | 11 | |
| TCGA | CNPY4 | 245812 | 52 | 6 | 38 |
Total number of gains: 133; Total number of losses: 25; Total Number of normals: 330.
Somatic mutations of CNPY4:
Generating mutation plots.
Highly correlated genes for CNPY4:
Showing top 20/290 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CNPY4 | EGR4 | 0.680955 | 3 | 0 | 3 |
| CNPY4 | KCTD17 | 0.675856 | 6 | 0 | 6 |
| CNPY4 | NPEPL1 | 0.662742 | 5 | 0 | 4 |
| CNPY4 | CHST8 | 0.657286 | 6 | 0 | 6 |
| CNPY4 | DIXDC1 | 0.652167 | 3 | 0 | 3 |
| CNPY4 | DHDH | 0.645921 | 3 | 0 | 3 |
| CNPY4 | MRM1 | 0.64401 | 3 | 0 | 3 |
| CNPY4 | TCF15 | 0.643606 | 5 | 0 | 5 |
| CNPY4 | FAM229B | 0.641626 | 3 | 0 | 3 |
| CNPY4 | USP27X-AS1 | 0.625741 | 4 | 0 | 3 |
| CNPY4 | ZHX3 | 0.622765 | 4 | 0 | 3 |
| CNPY4 | TRPC6 | 0.621533 | 3 | 0 | 3 |
| CNPY4 | PCBP4 | 0.619695 | 4 | 0 | 4 |
| CNPY4 | NEU2 | 0.617137 | 4 | 0 | 3 |
| CNPY4 | SGK2 | 0.615267 | 4 | 0 | 3 |
| CNPY4 | TMPRSS5 | 0.61494 | 3 | 0 | 3 |
| CNPY4 | TMEM263 | 0.614183 | 6 | 0 | 5 |
| CNPY4 | FAM131B | 0.607833 | 4 | 0 | 3 |
| CNPY4 | EID2B | 0.604458 | 5 | 0 | 4 |
| CNPY4 | DIO3 | 0.603865 | 4 | 0 | 3 |
For details and further investigation, click here