| Full name: kinase D interacting substrate 220 | Alias Symbol: ARMS | ||
| Type: protein-coding gene | Cytoband: 2p25.1 | ||
| Entrez ID: 57498 | HGNC ID: HGNC:29508 | Ensembl Gene: ENSG00000134313 | OMIM ID: 615759 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
KIDINS220 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04722 | Neurotrophin signaling pathway |
Expression of KIDINS220:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KIDINS220 | 57498 | 212163_at | -0.0241 | 0.9594 | |
| GSE20347 | KIDINS220 | 57498 | 212163_at | 0.2283 | 0.0753 | |
| GSE23400 | KIDINS220 | 57498 | 212163_at | 0.1377 | 0.1352 | |
| GSE26886 | KIDINS220 | 57498 | 212163_at | -0.0133 | 0.9613 | |
| GSE29001 | KIDINS220 | 57498 | 212163_at | 0.1208 | 0.7000 | |
| GSE38129 | KIDINS220 | 57498 | 212163_at | 0.1597 | 0.1580 | |
| GSE45670 | KIDINS220 | 57498 | 212163_at | -0.0536 | 0.6813 | |
| GSE53622 | KIDINS220 | 57498 | 104715 | 0.0557 | 0.4354 | |
| GSE53624 | KIDINS220 | 57498 | 130354 | -0.0337 | 0.6698 | |
| GSE63941 | KIDINS220 | 57498 | 212163_at | -1.4933 | 0.0011 | |
| GSE77861 | KIDINS220 | 57498 | 212163_at | 0.1801 | 0.3770 | |
| GSE97050 | KIDINS220 | 57498 | A_24_P7202 | -0.0387 | 0.8782 | |
| SRP007169 | KIDINS220 | 57498 | RNAseq | 0.5447 | 0.0662 | |
| SRP008496 | KIDINS220 | 57498 | RNAseq | 0.6543 | 0.0021 | |
| SRP064894 | KIDINS220 | 57498 | RNAseq | -0.2487 | 0.3779 | |
| SRP133303 | KIDINS220 | 57498 | RNAseq | 0.1125 | 0.2022 | |
| SRP159526 | KIDINS220 | 57498 | RNAseq | 0.2752 | 0.1481 | |
| SRP193095 | KIDINS220 | 57498 | RNAseq | 0.0279 | 0.7393 | |
| SRP219564 | KIDINS220 | 57498 | RNAseq | -0.0445 | 0.8418 | |
| TCGA | KIDINS220 | 57498 | RNAseq | -0.0133 | 0.7600 |
Upregulated datasets: 0; Downregulated datasets: 1.
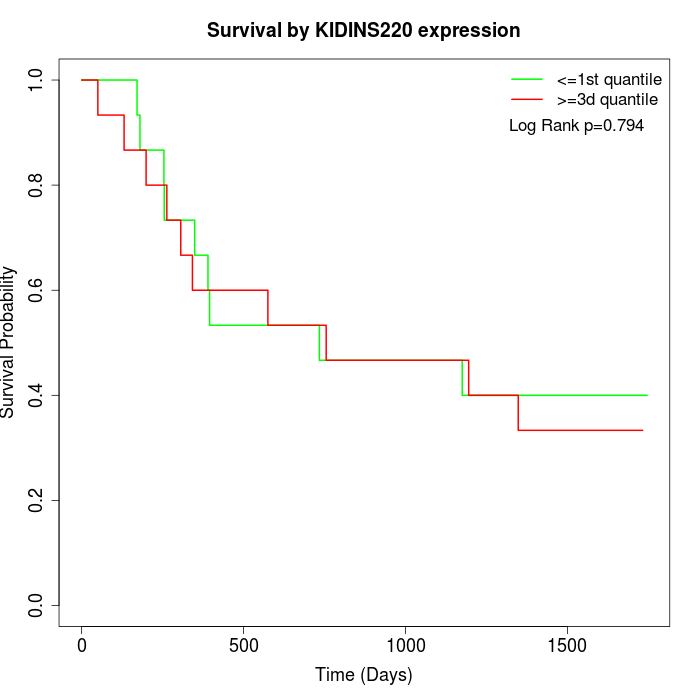
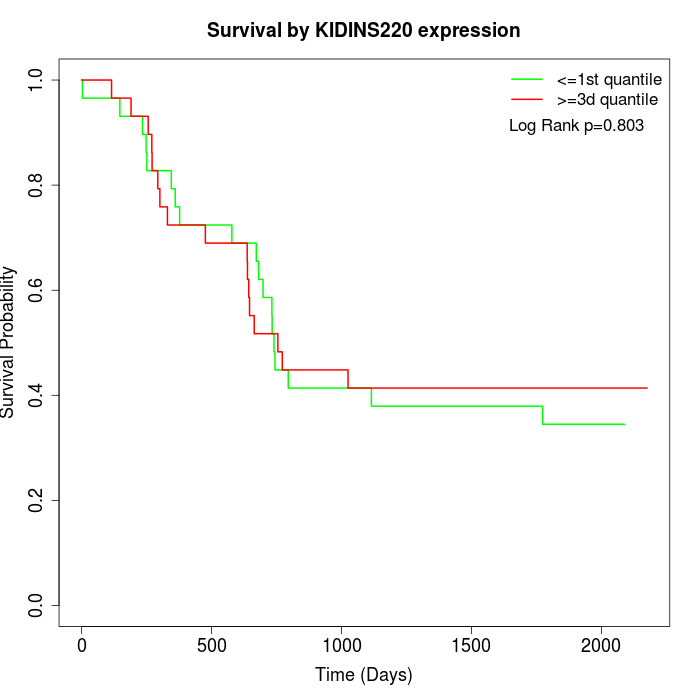
Survival by KIDINS220 expression:
Note: Click image to view full size file.
Copy number change of KIDINS220:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KIDINS220 | 57498 | 10 | 2 | 18 | |
| GSE20123 | KIDINS220 | 57498 | 10 | 2 | 18 | |
| GSE43470 | KIDINS220 | 57498 | 2 | 1 | 40 | |
| GSE46452 | KIDINS220 | 57498 | 3 | 4 | 52 | |
| GSE47630 | KIDINS220 | 57498 | 7 | 0 | 33 | |
| GSE54993 | KIDINS220 | 57498 | 0 | 7 | 63 | |
| GSE54994 | KIDINS220 | 57498 | 11 | 0 | 42 | |
| GSE60625 | KIDINS220 | 57498 | 0 | 3 | 8 | |
| GSE74703 | KIDINS220 | 57498 | 2 | 0 | 34 | |
| GSE74704 | KIDINS220 | 57498 | 9 | 1 | 10 | |
| TCGA | KIDINS220 | 57498 | 34 | 4 | 58 |
Total number of gains: 88; Total number of losses: 24; Total Number of normals: 376.
Somatic mutations of KIDINS220:
Generating mutation plots.
Highly correlated genes for KIDINS220:
Showing top 20/540 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KIDINS220 | EXOSC6 | 0.730686 | 3 | 0 | 3 |
| KIDINS220 | NDEL1 | 0.727161 | 3 | 0 | 3 |
| KIDINS220 | NAPG | 0.719222 | 3 | 0 | 3 |
| KIDINS220 | KBTBD6 | 0.714125 | 4 | 0 | 3 |
| KIDINS220 | DCTN6 | 0.7124 | 3 | 0 | 3 |
| KIDINS220 | WDR35 | 0.712211 | 3 | 0 | 3 |
| KIDINS220 | CHUK | 0.707373 | 3 | 0 | 3 |
| KIDINS220 | SIPA1L2 | 0.706313 | 3 | 0 | 3 |
| KIDINS220 | TRUB1 | 0.700136 | 3 | 0 | 3 |
| KIDINS220 | C16orf72 | 0.699698 | 3 | 0 | 3 |
| KIDINS220 | SLC35A1 | 0.6948 | 3 | 0 | 3 |
| KIDINS220 | HADHB | 0.693436 | 4 | 0 | 3 |
| KIDINS220 | TET1 | 0.686592 | 3 | 0 | 3 |
| KIDINS220 | IL13RA2 | 0.686249 | 3 | 0 | 3 |
| KIDINS220 | IPO8 | 0.685693 | 3 | 0 | 3 |
| KIDINS220 | ZSCAN29 | 0.684706 | 3 | 0 | 3 |
| KIDINS220 | FNIP2 | 0.683976 | 4 | 0 | 3 |
| KIDINS220 | ZFX | 0.678254 | 4 | 0 | 4 |
| KIDINS220 | CEP44 | 0.6776 | 4 | 0 | 4 |
| KIDINS220 | EIF2B4 | 0.677169 | 3 | 0 | 3 |
For details and further investigation, click here