| Full name: signal induced proliferation associated 1 like 2 | Alias Symbol: KIAA1389|SPAR2 | ||
| Type: protein-coding gene | Cytoband: 1q42.2 | ||
| Entrez ID: 57568 | HGNC ID: HGNC:23800 | Ensembl Gene: ENSG00000116991 | OMIM ID: 611609 |
| Drug and gene relationship at DGIdb | |||
SIPA1L2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04015 | Rap1 signaling pathway |
Expression of SIPA1L2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SIPA1L2 | 57568 | 225056_at | 0.2525 | 0.6343 | |
| GSE26886 | SIPA1L2 | 57568 | 225056_at | -0.1233 | 0.7643 | |
| GSE45670 | SIPA1L2 | 57568 | 225056_at | -0.2154 | 0.3725 | |
| GSE53622 | SIPA1L2 | 57568 | 76020 | -0.2638 | 0.0320 | |
| GSE53624 | SIPA1L2 | 57568 | 76020 | -0.3516 | 0.0012 | |
| GSE63941 | SIPA1L2 | 57568 | 225056_at | -0.4835 | 0.6869 | |
| GSE77861 | SIPA1L2 | 57568 | 225056_at | 0.3545 | 0.2210 | |
| GSE97050 | SIPA1L2 | 57568 | A_23_P137470 | -0.4282 | 0.2083 | |
| SRP007169 | SIPA1L2 | 57568 | RNAseq | -0.2345 | 0.5779 | |
| SRP008496 | SIPA1L2 | 57568 | RNAseq | 0.3667 | 0.2504 | |
| SRP064894 | SIPA1L2 | 57568 | RNAseq | -0.6581 | 0.0068 | |
| SRP133303 | SIPA1L2 | 57568 | RNAseq | -0.3283 | 0.1592 | |
| SRP159526 | SIPA1L2 | 57568 | RNAseq | 0.2874 | 0.6233 | |
| SRP193095 | SIPA1L2 | 57568 | RNAseq | -0.3241 | 0.1641 | |
| SRP219564 | SIPA1L2 | 57568 | RNAseq | -0.1409 | 0.5884 | |
| TCGA | SIPA1L2 | 57568 | RNAseq | 0.2318 | 0.0220 |
Upregulated datasets: 0; Downregulated datasets: 0.
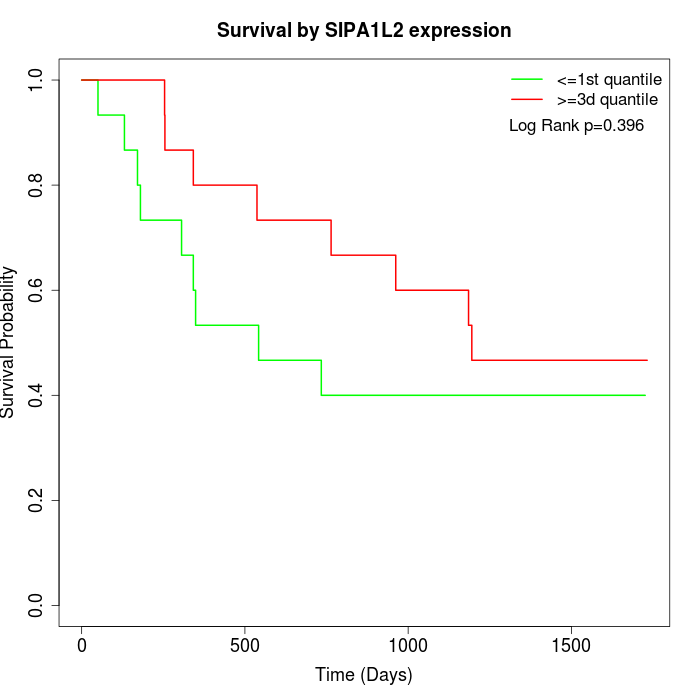
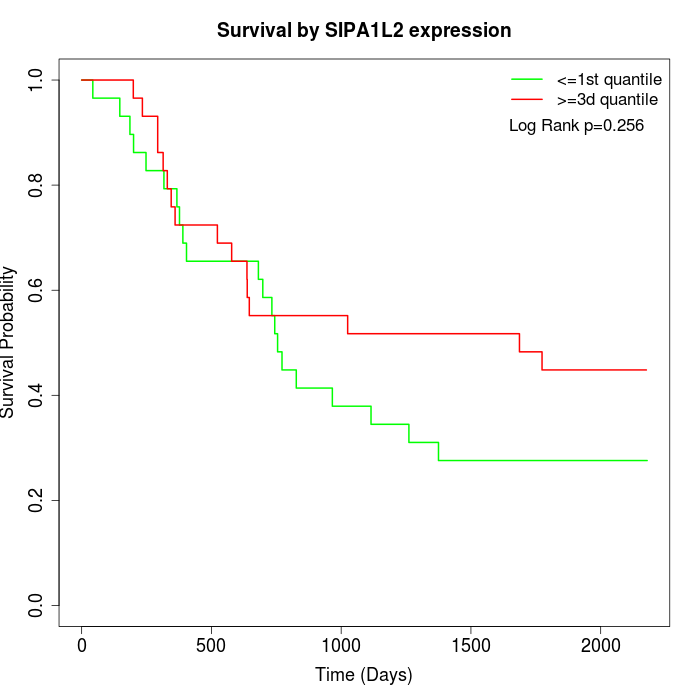
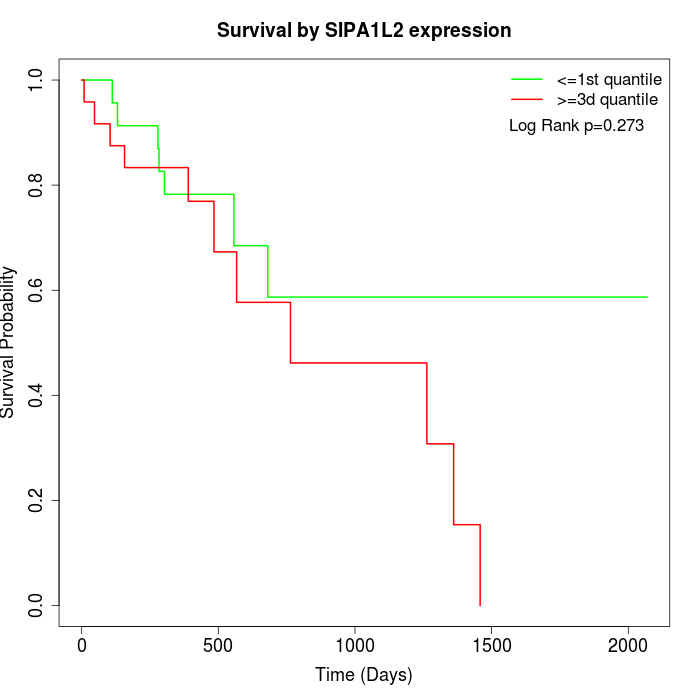
Survival by SIPA1L2 expression:
Note: Click image to view full size file.
Copy number change of SIPA1L2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SIPA1L2 | 57568 | 13 | 0 | 17 | |
| GSE20123 | SIPA1L2 | 57568 | 13 | 0 | 17 | |
| GSE43470 | SIPA1L2 | 57568 | 8 | 1 | 34 | |
| GSE46452 | SIPA1L2 | 57568 | 4 | 2 | 53 | |
| GSE47630 | SIPA1L2 | 57568 | 15 | 0 | 25 | |
| GSE54993 | SIPA1L2 | 57568 | 0 | 6 | 64 | |
| GSE54994 | SIPA1L2 | 57568 | 17 | 0 | 36 | |
| GSE60625 | SIPA1L2 | 57568 | 0 | 0 | 11 | |
| GSE74703 | SIPA1L2 | 57568 | 8 | 1 | 27 | |
| GSE74704 | SIPA1L2 | 57568 | 7 | 0 | 13 | |
| TCGA | SIPA1L2 | 57568 | 44 | 3 | 49 |
Total number of gains: 129; Total number of losses: 13; Total Number of normals: 346.
Somatic mutations of SIPA1L2:
Generating mutation plots.
Highly correlated genes for SIPA1L2:
Showing top 20/123 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SIPA1L2 | BRD4 | 0.849341 | 3 | 0 | 3 |
| SIPA1L2 | ULK1 | 0.798417 | 3 | 0 | 3 |
| SIPA1L2 | ERGIC2 | 0.796394 | 3 | 0 | 3 |
| SIPA1L2 | SCAF8 | 0.793142 | 3 | 0 | 3 |
| SIPA1L2 | SURF6 | 0.784918 | 3 | 0 | 3 |
| SIPA1L2 | SUZ12 | 0.769246 | 3 | 0 | 3 |
| SIPA1L2 | GNA11 | 0.768752 | 3 | 0 | 3 |
| SIPA1L2 | MTRF1 | 0.75878 | 3 | 0 | 3 |
| SIPA1L2 | LUC7L3 | 0.755014 | 3 | 0 | 3 |
| SIPA1L2 | FASTKD1 | 0.7536 | 3 | 0 | 3 |
| SIPA1L2 | RAB3GAP1 | 0.751459 | 4 | 0 | 4 |
| SIPA1L2 | HELZ | 0.750541 | 3 | 0 | 3 |
| SIPA1L2 | MYNN | 0.74274 | 3 | 0 | 3 |
| SIPA1L2 | TANC1 | 0.740904 | 4 | 0 | 4 |
| SIPA1L2 | GOSR1 | 0.737673 | 4 | 0 | 4 |
| SIPA1L2 | KPNA1 | 0.735198 | 3 | 0 | 3 |
| SIPA1L2 | CHD8 | 0.7287 | 3 | 0 | 3 |
| SIPA1L2 | ZNF780B | 0.726151 | 3 | 0 | 3 |
| SIPA1L2 | CCDC93 | 0.724402 | 4 | 0 | 4 |
| SIPA1L2 | UPF1 | 0.723429 | 3 | 0 | 3 |
For details and further investigation, click here