| Full name: kelch like family member 12 | Alias Symbol: C3IP1 | ||
| Type: protein-coding gene | Cytoband: 1q32.1 | ||
| Entrez ID: 59349 | HGNC ID: HGNC:19360 | Ensembl Gene: ENSG00000117153 | OMIM ID: 614522 |
| Drug and gene relationship at DGIdb | |||
Expression of KLHL12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KLHL12 | 59349 | 225068_at | 0.0266 | 0.9675 | |
| GSE20347 | KLHL12 | 59349 | 219931_s_at | 0.0289 | 0.8558 | |
| GSE23400 | KLHL12 | 59349 | 219931_s_at | 0.0632 | 0.1693 | |
| GSE26886 | KLHL12 | 59349 | 225068_at | -0.0215 | 0.9432 | |
| GSE29001 | KLHL12 | 59349 | 219931_s_at | -0.3558 | 0.1182 | |
| GSE38129 | KLHL12 | 59349 | 219931_s_at | 0.2153 | 0.0963 | |
| GSE45670 | KLHL12 | 59349 | 225068_at | 0.1965 | 0.1995 | |
| GSE53622 | KLHL12 | 59349 | 24879 | 0.4456 | 0.0000 | |
| GSE53624 | KLHL12 | 59349 | 24879 | 0.3925 | 0.0000 | |
| GSE63941 | KLHL12 | 59349 | 225068_at | 0.5987 | 0.1025 | |
| GSE77861 | KLHL12 | 59349 | 225068_at | 0.2766 | 0.1761 | |
| GSE97050 | KLHL12 | 59349 | A_33_P3394021 | 0.3651 | 0.2464 | |
| SRP007169 | KLHL12 | 59349 | RNAseq | -0.2699 | 0.3646 | |
| SRP008496 | KLHL12 | 59349 | RNAseq | -0.2118 | 0.3645 | |
| SRP064894 | KLHL12 | 59349 | RNAseq | -0.0255 | 0.8389 | |
| SRP133303 | KLHL12 | 59349 | RNAseq | 0.3680 | 0.0104 | |
| SRP159526 | KLHL12 | 59349 | RNAseq | 0.3293 | 0.0673 | |
| SRP193095 | KLHL12 | 59349 | RNAseq | -0.0159 | 0.8473 | |
| SRP219564 | KLHL12 | 59349 | RNAseq | 0.1578 | 0.5143 | |
| TCGA | KLHL12 | 59349 | RNAseq | 0.0691 | 0.1784 |
Upregulated datasets: 0; Downregulated datasets: 0.
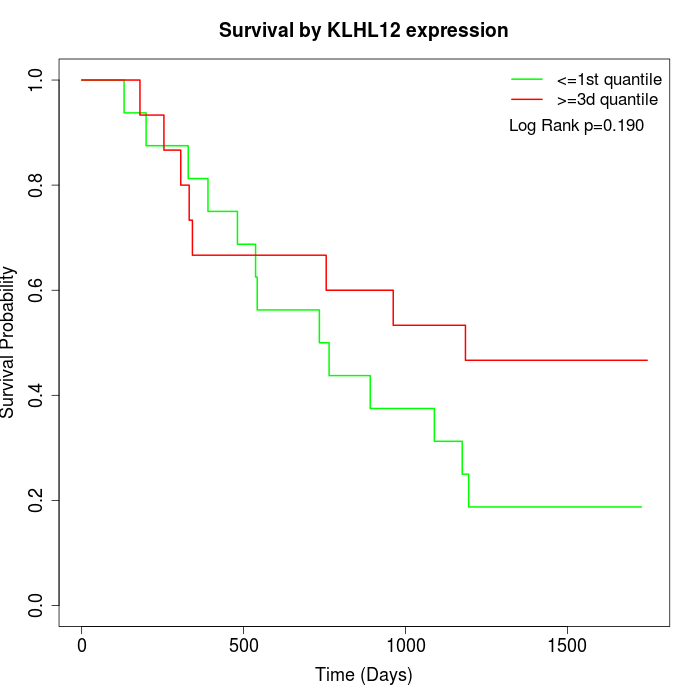
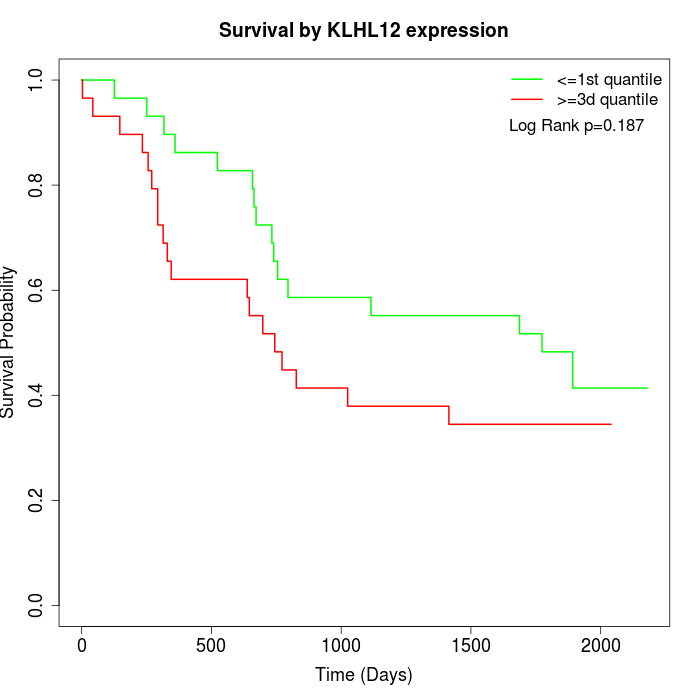
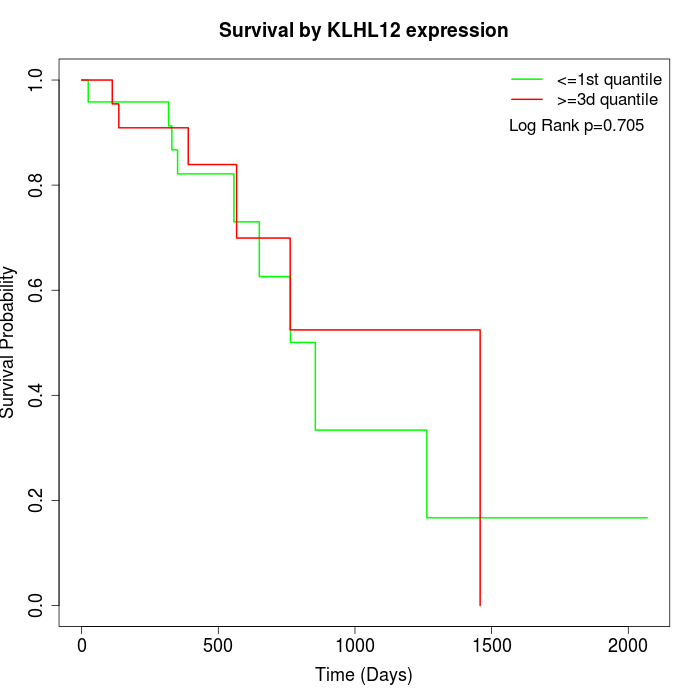
Survival by KLHL12 expression:
Note: Click image to view full size file.
Copy number change of KLHL12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KLHL12 | 59349 | 11 | 0 | 19 | |
| GSE20123 | KLHL12 | 59349 | 11 | 0 | 19 | |
| GSE43470 | KLHL12 | 59349 | 6 | 0 | 37 | |
| GSE46452 | KLHL12 | 59349 | 3 | 1 | 55 | |
| GSE47630 | KLHL12 | 59349 | 14 | 0 | 26 | |
| GSE54993 | KLHL12 | 59349 | 0 | 6 | 64 | |
| GSE54994 | KLHL12 | 59349 | 15 | 0 | 38 | |
| GSE60625 | KLHL12 | 59349 | 0 | 0 | 11 | |
| GSE74703 | KLHL12 | 59349 | 6 | 0 | 30 | |
| GSE74704 | KLHL12 | 59349 | 5 | 0 | 15 | |
| TCGA | KLHL12 | 59349 | 44 | 4 | 48 |
Total number of gains: 115; Total number of losses: 11; Total Number of normals: 362.
Somatic mutations of KLHL12:
Generating mutation plots.
Highly correlated genes for KLHL12:
Showing top 20/655 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KLHL12 | MMS22L | 0.70816 | 3 | 0 | 3 |
| KLHL12 | LMBRD2 | 0.700865 | 3 | 0 | 3 |
| KLHL12 | ATM | 0.694727 | 3 | 0 | 3 |
| KLHL12 | GPANK1 | 0.682775 | 3 | 0 | 3 |
| KLHL12 | WDR54 | 0.673422 | 4 | 0 | 4 |
| KLHL12 | CTR9 | 0.667261 | 4 | 0 | 3 |
| KLHL12 | SEC11C | 0.66356 | 4 | 0 | 3 |
| KLHL12 | SNAP47 | 0.661986 | 4 | 0 | 3 |
| KLHL12 | GMCL1 | 0.655735 | 5 | 0 | 5 |
| KLHL12 | IER3IP1 | 0.654157 | 4 | 0 | 3 |
| KLHL12 | PPP1R8 | 0.654009 | 3 | 0 | 3 |
| KLHL12 | B3GALT6 | 0.653851 | 3 | 0 | 3 |
| KLHL12 | BROX | 0.653415 | 3 | 0 | 3 |
| KLHL12 | PLEKHF2 | 0.651986 | 3 | 0 | 3 |
| KLHL12 | TBC1D15 | 0.651279 | 4 | 0 | 4 |
| KLHL12 | RPL26L1 | 0.650688 | 3 | 0 | 3 |
| KLHL12 | MAGEA6 | 0.650054 | 4 | 0 | 3 |
| KLHL12 | SLC39A11 | 0.649377 | 3 | 0 | 3 |
| KLHL12 | PMS2 | 0.646945 | 4 | 0 | 4 |
| KLHL12 | DYNLRB1 | 0.641803 | 3 | 0 | 3 |
For details and further investigation, click here