| Full name: kinectin 1 | Alias Symbol: KIAA0004|CG1|KNT | ||
| Type: protein-coding gene | Cytoband: 14q22.1 | ||
| Entrez ID: 3895 | HGNC ID: HGNC:6467 | Ensembl Gene: ENSG00000126777 | OMIM ID: 600381 |
| Drug and gene relationship at DGIdb | |||
Expression of KTN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KTN1 | 3895 | 200915_x_at | 0.2149 | 0.6192 | |
| GSE20347 | KTN1 | 3895 | 200915_x_at | 0.1749 | 0.2897 | |
| GSE23400 | KTN1 | 3895 | 200915_x_at | 0.6223 | 0.0000 | |
| GSE26886 | KTN1 | 3895 | 200915_x_at | 1.2022 | 0.0000 | |
| GSE29001 | KTN1 | 3895 | 200915_x_at | 0.2731 | 0.3733 | |
| GSE38129 | KTN1 | 3895 | 200915_x_at | 0.1646 | 0.1387 | |
| GSE45670 | KTN1 | 3895 | 200915_x_at | 0.2094 | 0.2074 | |
| GSE53622 | KTN1 | 3895 | 43286 | 0.5296 | 0.0000 | |
| GSE53624 | KTN1 | 3895 | 43286 | 0.4882 | 0.0000 | |
| GSE63941 | KTN1 | 3895 | 200915_x_at | -0.1288 | 0.8216 | |
| GSE77861 | KTN1 | 3895 | 200915_x_at | 0.5648 | 0.1869 | |
| GSE97050 | KTN1 | 3895 | A_33_P3348639 | 0.2811 | 0.3379 | |
| SRP007169 | KTN1 | 3895 | RNAseq | 1.2724 | 0.0114 | |
| SRP008496 | KTN1 | 3895 | RNAseq | 1.4363 | 0.0000 | |
| SRP064894 | KTN1 | 3895 | RNAseq | 0.2843 | 0.1015 | |
| SRP133303 | KTN1 | 3895 | RNAseq | 0.4881 | 0.0013 | |
| SRP159526 | KTN1 | 3895 | RNAseq | 0.3505 | 0.3127 | |
| SRP193095 | KTN1 | 3895 | RNAseq | 0.4022 | 0.0305 | |
| SRP219564 | KTN1 | 3895 | RNAseq | 0.1431 | 0.7223 | |
| TCGA | KTN1 | 3895 | RNAseq | 0.0846 | 0.0887 |
Upregulated datasets: 3; Downregulated datasets: 0.
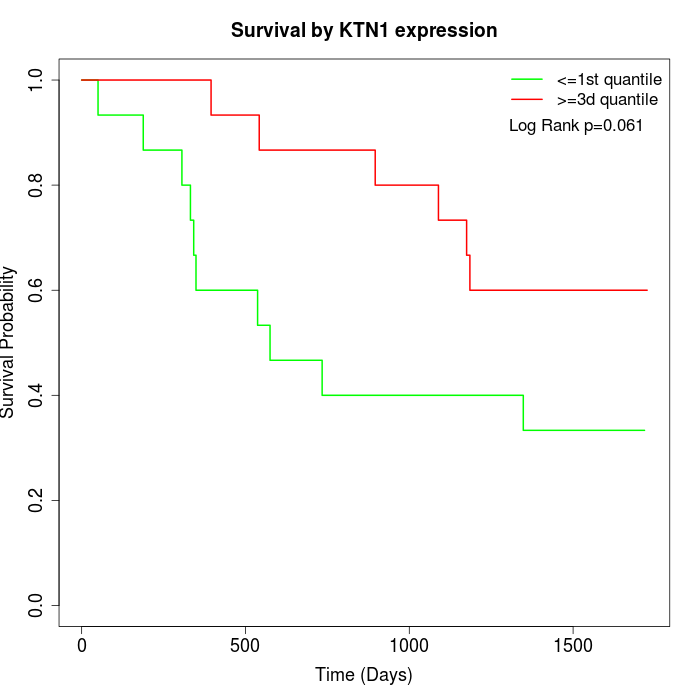
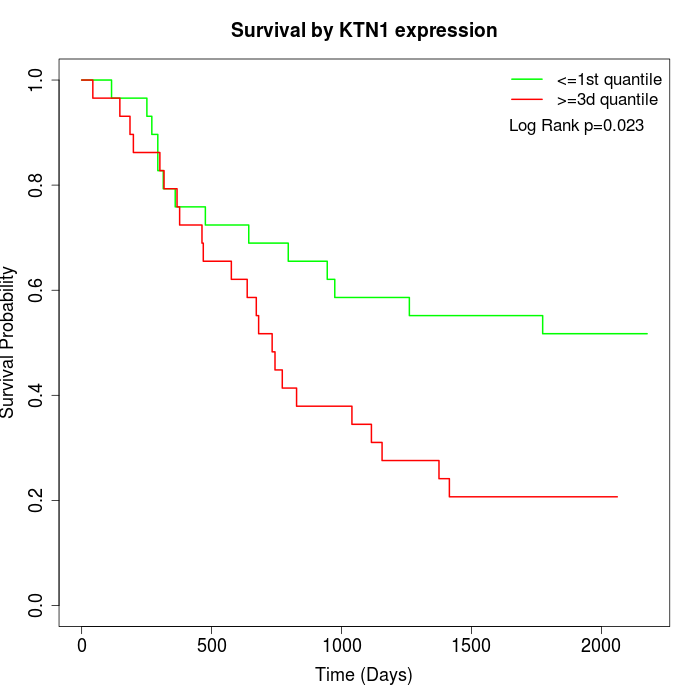
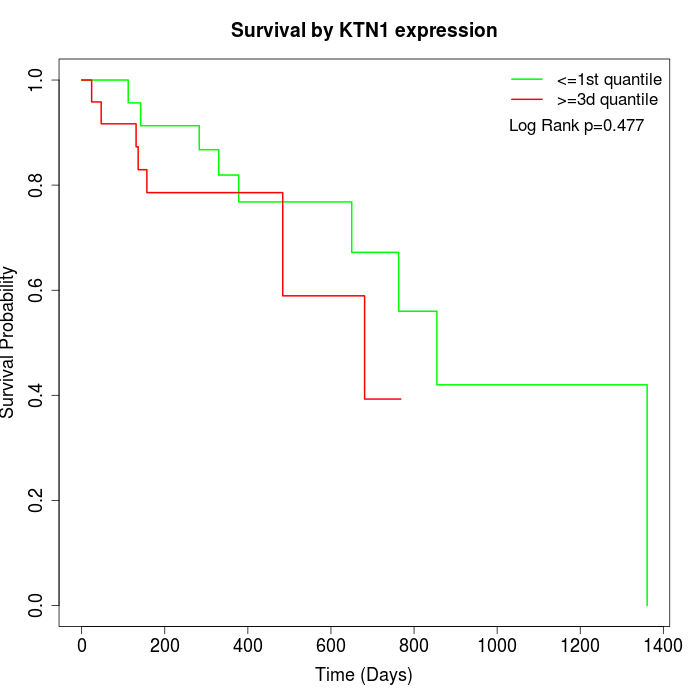
Survival by KTN1 expression:
Note: Click image to view full size file.
Copy number change of KTN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KTN1 | 3895 | 8 | 3 | 19 | |
| GSE20123 | KTN1 | 3895 | 8 | 3 | 19 | |
| GSE43470 | KTN1 | 3895 | 8 | 1 | 34 | |
| GSE46452 | KTN1 | 3895 | 16 | 3 | 40 | |
| GSE47630 | KTN1 | 3895 | 11 | 10 | 19 | |
| GSE54993 | KTN1 | 3895 | 3 | 8 | 59 | |
| GSE54994 | KTN1 | 3895 | 19 | 4 | 30 | |
| GSE60625 | KTN1 | 3895 | 0 | 2 | 9 | |
| GSE74703 | KTN1 | 3895 | 7 | 1 | 28 | |
| GSE74704 | KTN1 | 3895 | 3 | 2 | 15 | |
| TCGA | KTN1 | 3895 | 32 | 14 | 50 |
Total number of gains: 115; Total number of losses: 51; Total Number of normals: 322.
Somatic mutations of KTN1:
Generating mutation plots.
Highly correlated genes for KTN1:
Showing top 20/1323 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KTN1 | THEM4 | 0.862698 | 3 | 0 | 3 |
| KTN1 | PET117 | 0.853912 | 3 | 0 | 3 |
| KTN1 | TIMM9 | 0.762253 | 12 | 0 | 12 |
| KTN1 | MNAT1 | 0.755411 | 12 | 0 | 12 |
| KTN1 | FERMT2 | 0.751079 | 3 | 0 | 3 |
| KTN1 | SNW1 | 0.747182 | 10 | 0 | 10 |
| KTN1 | FARS2 | 0.745408 | 3 | 0 | 3 |
| KTN1 | MRPS5 | 0.739918 | 3 | 0 | 3 |
| KTN1 | COCH | 0.731243 | 5 | 0 | 5 |
| KTN1 | WDR54 | 0.726595 | 6 | 0 | 6 |
| KTN1 | SCAMP1 | 0.725764 | 3 | 0 | 3 |
| KTN1 | SOCS4 | 0.718132 | 9 | 0 | 9 |
| KTN1 | UBTD2 | 0.709967 | 7 | 0 | 7 |
| KTN1 | CEP63 | 0.709896 | 4 | 0 | 4 |
| KTN1 | RNMT | 0.703577 | 4 | 0 | 4 |
| KTN1 | SMARCAL1 | 0.702023 | 3 | 0 | 3 |
| KTN1 | UAP1 | 0.701495 | 3 | 0 | 3 |
| KTN1 | NPHP3 | 0.693605 | 4 | 0 | 3 |
| KTN1 | IFT43 | 0.691171 | 6 | 0 | 6 |
| KTN1 | CENPU | 0.687365 | 3 | 0 | 3 |
For details and further investigation, click here