| Full name: SNW domain containing 1 | Alias Symbol: NCoA-62|SKIP|Prp45|PRPF45|Bx42|SKIP1|FUN20 | ||
| Type: protein-coding gene | Cytoband: 14q24.3 | ||
| Entrez ID: 22938 | HGNC ID: HGNC:16696 | Ensembl Gene: ENSG00000100603 | OMIM ID: 603055 |
| Drug and gene relationship at DGIdb | |||
SNW1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04330 | Notch signaling pathway |
Expression of SNW1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SNW1 | 22938 | 201575_at | 0.0550 | 0.9063 | |
| GSE20347 | SNW1 | 22938 | 201575_at | 0.3590 | 0.0013 | |
| GSE23400 | SNW1 | 22938 | 201575_at | 0.5556 | 0.0000 | |
| GSE26886 | SNW1 | 22938 | 201575_at | 0.6094 | 0.0030 | |
| GSE29001 | SNW1 | 22938 | 201575_at | 0.5027 | 0.0156 | |
| GSE38129 | SNW1 | 22938 | 201575_at | 0.3312 | 0.0000 | |
| GSE45670 | SNW1 | 22938 | 201575_at | 0.2420 | 0.0480 | |
| GSE53622 | SNW1 | 22938 | 10374 | 0.3743 | 0.0000 | |
| GSE53624 | SNW1 | 22938 | 10374 | 0.4883 | 0.0000 | |
| GSE63941 | SNW1 | 22938 | 201575_at | 0.7392 | 0.0048 | |
| GSE77861 | SNW1 | 22938 | 201575_at | 0.3945 | 0.1422 | |
| GSE97050 | SNW1 | 22938 | A_23_P37347 | 0.1837 | 0.5058 | |
| SRP007169 | SNW1 | 22938 | RNAseq | 1.3024 | 0.0043 | |
| SRP008496 | SNW1 | 22938 | RNAseq | 1.4394 | 0.0000 | |
| SRP064894 | SNW1 | 22938 | RNAseq | 0.4041 | 0.0096 | |
| SRP133303 | SNW1 | 22938 | RNAseq | 0.4544 | 0.0010 | |
| SRP159526 | SNW1 | 22938 | RNAseq | 0.1734 | 0.4964 | |
| SRP193095 | SNW1 | 22938 | RNAseq | 0.2299 | 0.0328 | |
| SRP219564 | SNW1 | 22938 | RNAseq | 0.2624 | 0.4449 | |
| TCGA | SNW1 | 22938 | RNAseq | 0.1378 | 0.0053 |
Upregulated datasets: 2; Downregulated datasets: 0.
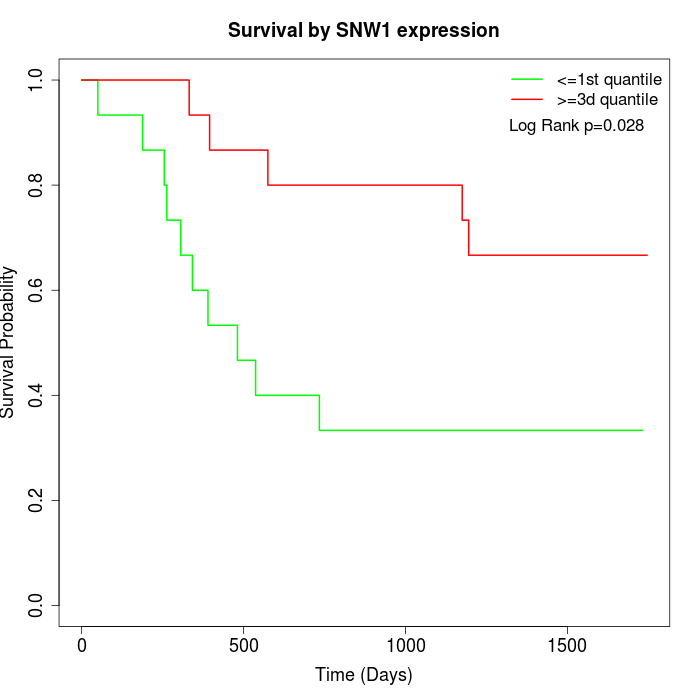
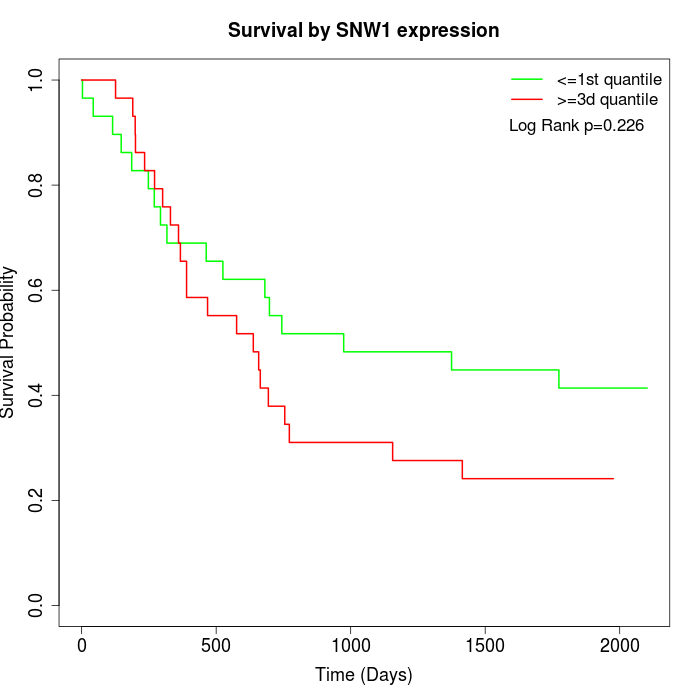
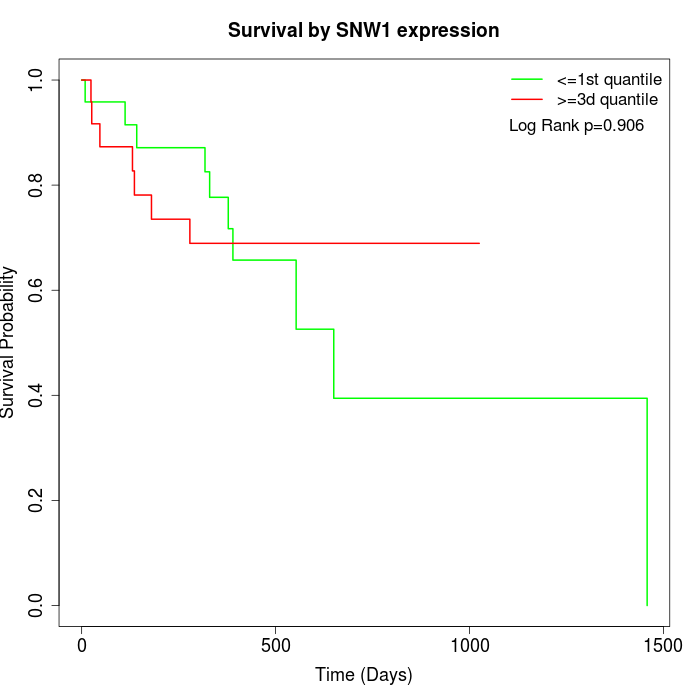
Survival by SNW1 expression:
Note: Click image to view full size file.
Copy number change of SNW1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SNW1 | 22938 | 7 | 3 | 20 | |
| GSE20123 | SNW1 | 22938 | 7 | 3 | 20 | |
| GSE43470 | SNW1 | 22938 | 8 | 3 | 32 | |
| GSE46452 | SNW1 | 22938 | 16 | 3 | 40 | |
| GSE47630 | SNW1 | 22938 | 11 | 8 | 21 | |
| GSE54993 | SNW1 | 22938 | 3 | 8 | 59 | |
| GSE54994 | SNW1 | 22938 | 19 | 4 | 30 | |
| GSE60625 | SNW1 | 22938 | 0 | 2 | 9 | |
| GSE74703 | SNW1 | 22938 | 7 | 3 | 26 | |
| GSE74704 | SNW1 | 22938 | 3 | 3 | 14 | |
| TCGA | SNW1 | 22938 | 31 | 16 | 49 |
Total number of gains: 112; Total number of losses: 56; Total Number of normals: 320.
Somatic mutations of SNW1:
Generating mutation plots.
Highly correlated genes for SNW1:
Showing top 20/1127 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SNW1 | RAD17 | 0.770931 | 3 | 0 | 3 |
| SNW1 | ATP11C | 0.770917 | 3 | 0 | 3 |
| SNW1 | ANKRD10 | 0.770425 | 3 | 0 | 3 |
| SNW1 | JKAMP | 0.754185 | 6 | 0 | 6 |
| SNW1 | KTN1 | 0.747182 | 10 | 0 | 10 |
| SNW1 | SPIRE1 | 0.74102 | 3 | 0 | 3 |
| SNW1 | ALKBH1 | 0.737351 | 12 | 0 | 11 |
| SNW1 | PET117 | 0.732785 | 4 | 0 | 3 |
| SNW1 | IRX3 | 0.730117 | 3 | 0 | 3 |
| SNW1 | STAG1 | 0.729868 | 3 | 0 | 3 |
| SNW1 | KLF13 | 0.727655 | 3 | 0 | 3 |
| SNW1 | CDC42SE2 | 0.726903 | 4 | 0 | 4 |
| SNW1 | CBFA2T2 | 0.723277 | 3 | 0 | 3 |
| SNW1 | BOD1 | 0.717977 | 6 | 0 | 6 |
| SNW1 | CELSR2 | 0.71521 | 3 | 0 | 3 |
| SNW1 | TOX4 | 0.712402 | 4 | 0 | 4 |
| SNW1 | PACSIN2 | 0.712336 | 3 | 0 | 3 |
| SNW1 | TCF7L2 | 0.711165 | 3 | 0 | 3 |
| SNW1 | WIPI2 | 0.70966 | 3 | 0 | 3 |
| SNW1 | SFMBT1 | 0.709312 | 3 | 0 | 3 |
For details and further investigation, click here