| Full name: laminin subunit beta 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 7q31.1 | ||
| Entrez ID: 3912 | HGNC ID: HGNC:6486 | Ensembl Gene: ENSG00000091136 | OMIM ID: 150240 |
| Drug and gene relationship at DGIdb | |||
LAMB1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04510 | Focal adhesion | |
| hsa05200 | Pathways in cancer | |
| hsa05222 | Small cell lung cancer |
Expression of LAMB1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LAMB1 | 3912 | 201505_at | 0.4288 | 0.6344 | |
| GSE20347 | LAMB1 | 3912 | 201505_at | 1.3333 | 0.0000 | |
| GSE23400 | LAMB1 | 3912 | 211651_s_at | 0.9782 | 0.0000 | |
| GSE26886 | LAMB1 | 3912 | 201505_at | 2.9580 | 0.0000 | |
| GSE29001 | LAMB1 | 3912 | 201505_at | 1.1562 | 0.0227 | |
| GSE38129 | LAMB1 | 3912 | 201505_at | 1.1763 | 0.0000 | |
| GSE45670 | LAMB1 | 3912 | 201505_at | 0.2279 | 0.5245 | |
| GSE53622 | LAMB1 | 3912 | 6055 | 1.0495 | 0.0000 | |
| GSE53624 | LAMB1 | 3912 | 6055 | 1.2145 | 0.0000 | |
| GSE63941 | LAMB1 | 3912 | 211651_s_at | -4.3500 | 0.0007 | |
| GSE77861 | LAMB1 | 3912 | 211651_s_at | 0.8814 | 0.0629 | |
| GSE97050 | LAMB1 | 3912 | A_23_P94030 | 0.6548 | 0.1194 | |
| SRP007169 | LAMB1 | 3912 | RNAseq | 3.3077 | 0.0000 | |
| SRP008496 | LAMB1 | 3912 | RNAseq | 3.0778 | 0.0000 | |
| SRP064894 | LAMB1 | 3912 | RNAseq | 1.5233 | 0.0000 | |
| SRP133303 | LAMB1 | 3912 | RNAseq | 0.8781 | 0.0028 | |
| SRP159526 | LAMB1 | 3912 | RNAseq | 1.2539 | 0.0022 | |
| SRP193095 | LAMB1 | 3912 | RNAseq | 1.3157 | 0.0000 | |
| SRP219564 | LAMB1 | 3912 | RNAseq | 0.7537 | 0.2091 | |
| TCGA | LAMB1 | 3912 | RNAseq | -0.0106 | 0.8876 |
Upregulated datasets: 11; Downregulated datasets: 1.
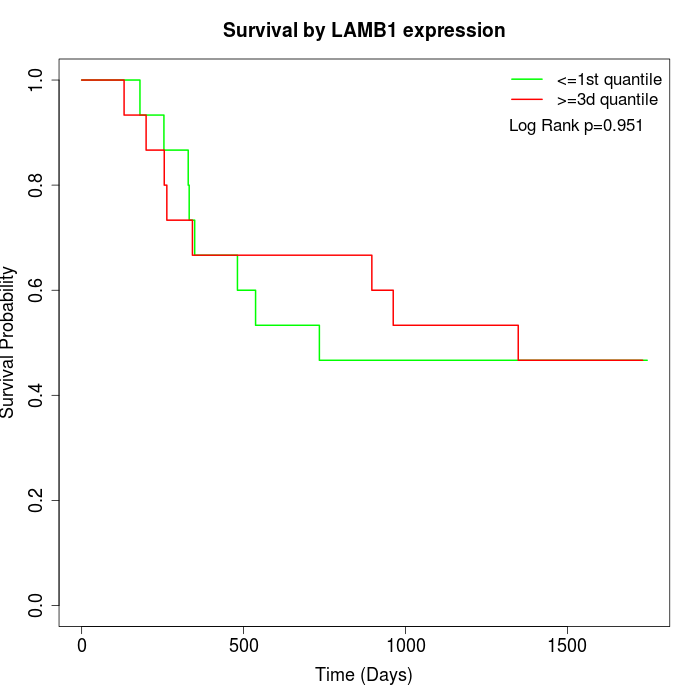
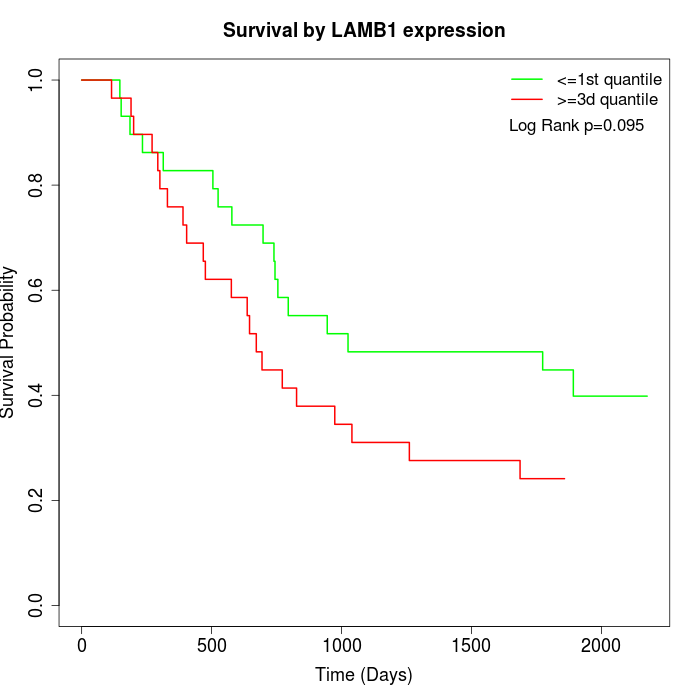
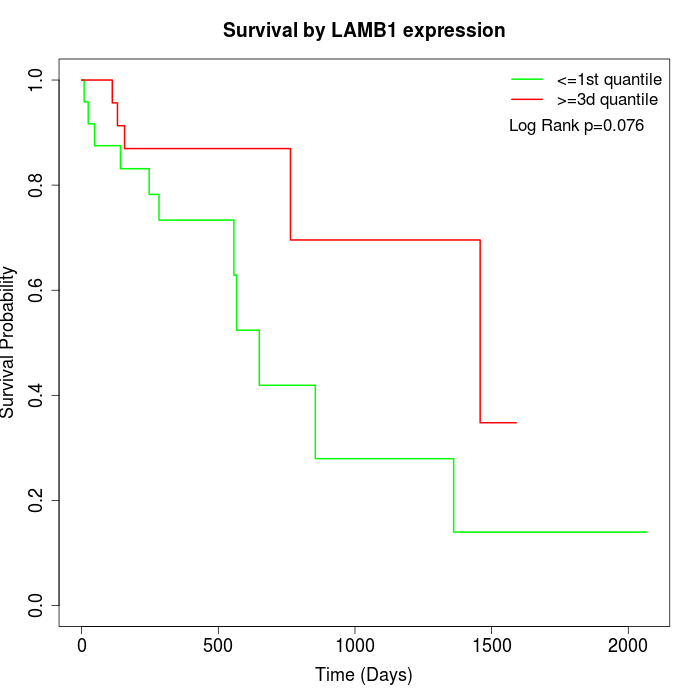
Survival by LAMB1 expression:
Note: Click image to view full size file.
Copy number change of LAMB1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LAMB1 | 3912 | 11 | 0 | 19 | |
| GSE20123 | LAMB1 | 3912 | 11 | 0 | 19 | |
| GSE43470 | LAMB1 | 3912 | 6 | 2 | 35 | |
| GSE46452 | LAMB1 | 3912 | 10 | 1 | 48 | |
| GSE47630 | LAMB1 | 3912 | 8 | 3 | 29 | |
| GSE54993 | LAMB1 | 3912 | 2 | 9 | 59 | |
| GSE54994 | LAMB1 | 3912 | 16 | 3 | 34 | |
| GSE60625 | LAMB1 | 3912 | 0 | 0 | 11 | |
| GSE74703 | LAMB1 | 3912 | 6 | 2 | 28 | |
| GSE74704 | LAMB1 | 3912 | 7 | 0 | 13 | |
| TCGA | LAMB1 | 3912 | 51 | 10 | 35 |
Total number of gains: 128; Total number of losses: 30; Total Number of normals: 330.
Somatic mutations of LAMB1:
Generating mutation plots.
Highly correlated genes for LAMB1:
Showing top 20/1460 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LAMB1 | RAB34 | 0.810156 | 3 | 0 | 3 |
| LAMB1 | FADS1 | 0.789815 | 3 | 0 | 3 |
| LAMB1 | TMEM263 | 0.765489 | 6 | 0 | 6 |
| LAMB1 | COL4A1 | 0.762904 | 13 | 0 | 13 |
| LAMB1 | GNG10 | 0.762158 | 3 | 0 | 3 |
| LAMB1 | FKBP9 | 0.761095 | 10 | 0 | 9 |
| LAMB1 | RAI14 | 0.757627 | 11 | 0 | 10 |
| LAMB1 | CCDC127 | 0.755287 | 3 | 0 | 3 |
| LAMB1 | CEP170 | 0.754298 | 3 | 0 | 3 |
| LAMB1 | LAMC1 | 0.750165 | 13 | 0 | 13 |
| LAMB1 | LOXL2 | 0.74822 | 11 | 0 | 11 |
| LAMB1 | CDH11 | 0.747551 | 13 | 0 | 12 |
| LAMB1 | VCAN | 0.742956 | 13 | 0 | 12 |
| LAMB1 | FSTL1 | 0.740808 | 12 | 0 | 12 |
| LAMB1 | COL3A1 | 0.739266 | 11 | 0 | 11 |
| LAMB1 | FAM160B1 | 0.7373 | 3 | 0 | 3 |
| LAMB1 | GXYLT2 | 0.736743 | 7 | 0 | 7 |
| LAMB1 | GPX8 | 0.734466 | 8 | 0 | 7 |
| LAMB1 | SPARC | 0.733537 | 13 | 0 | 12 |
| LAMB1 | PSME1 | 0.731904 | 3 | 0 | 3 |
For details and further investigation, click here