| Full name: centrosomal protein 170 | Alias Symbol: KAB|FAM68A | ||
| Type: protein-coding gene | Cytoband: 1q43 | ||
| Entrez ID: 9859 | HGNC ID: HGNC:28920 | Ensembl Gene: ENSG00000143702 | OMIM ID: 613023 |
| Drug and gene relationship at DGIdb | |||
Expression of CEP170:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | CEP170 | 9859 | 86215 | 0.8075 | 0.0000 | |
| GSE53624 | CEP170 | 9859 | 58260 | 0.8394 | 0.0000 | |
| GSE97050 | CEP170 | A_21_P0012605 | 0.1619 | 0.7312 | ||
| SRP007169 | CEP170 | 9859 | RNAseq | 1.9821 | 0.0000 | |
| SRP008496 | CEP170 | 9859 | RNAseq | 1.9282 | 0.0000 | |
| SRP064894 | CEP170 | 9859 | RNAseq | 0.8648 | 0.0056 | |
| SRP133303 | CEP170 | 9859 | RNAseq | 0.7342 | 0.0000 | |
| SRP159526 | CEP170 | 9859 | RNAseq | 0.6760 | 0.0128 | |
| SRP193095 | CEP170 | 9859 | RNAseq | 0.9605 | 0.0000 | |
| SRP219564 | CEP170 | 9859 | RNAseq | 0.7026 | 0.0061 | |
| TCGA | CEP170 | 9859 | RNAseq | 0.2470 | 0.0003 |
Upregulated datasets: 2; Downregulated datasets: 0.
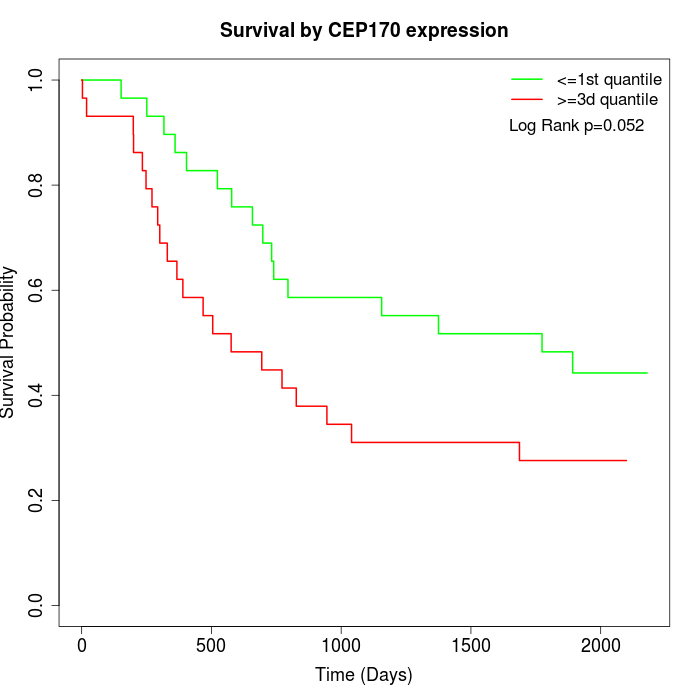
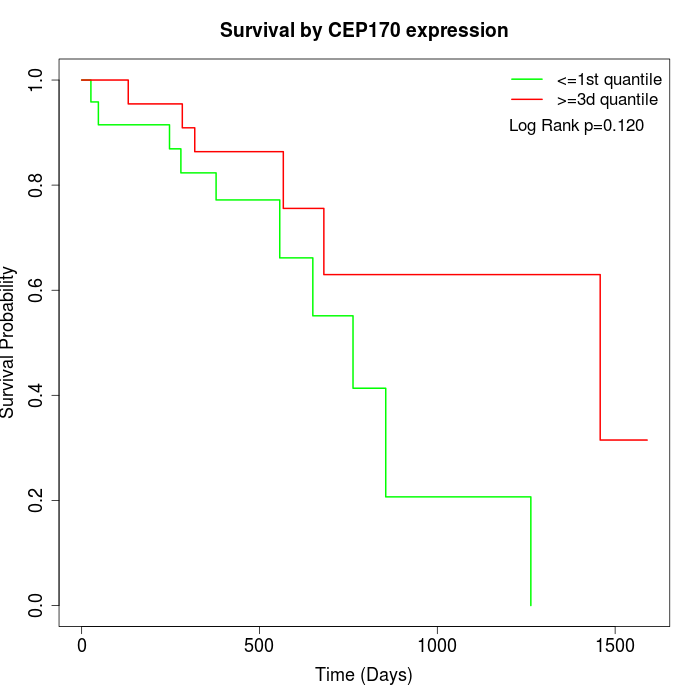
Survival by CEP170 expression:
Note: Click image to view full size file.
Copy number change of CEP170:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CEP170 | 9859 | 7 | 1 | 22 | |
| GSE20123 | CEP170 | 9859 | 7 | 1 | 22 | |
| GSE43470 | CEP170 | 9859 | 6 | 2 | 35 | |
| GSE46452 | CEP170 | 9859 | 4 | 2 | 53 | |
| GSE47630 | CEP170 | 9859 | 15 | 0 | 25 | |
| GSE54993 | CEP170 | 9859 | 0 | 6 | 64 | |
| GSE54994 | CEP170 | 9859 | 18 | 0 | 35 | |
| GSE60625 | CEP170 | 9859 | 0 | 0 | 11 | |
| GSE74703 | CEP170 | 9859 | 6 | 1 | 29 | |
| GSE74704 | CEP170 | 9859 | 2 | 0 | 18 | |
| TCGA | CEP170 | 9859 | 45 | 6 | 45 |
Total number of gains: 110; Total number of losses: 19; Total Number of normals: 359.
Somatic mutations of CEP170:
Generating mutation plots.
Highly correlated genes for CEP170:
Showing top 20/407 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CEP170 | GNA12 | 0.802579 | 3 | 0 | 3 |
| CEP170 | GPR137B | 0.78713 | 3 | 0 | 3 |
| CEP170 | SLC39A10 | 0.773882 | 3 | 0 | 3 |
| CEP170 | RAI14 | 0.767506 | 3 | 0 | 3 |
| CEP170 | ZNF281 | 0.765943 | 3 | 0 | 3 |
| CEP170 | FZD2 | 0.761629 | 3 | 0 | 3 |
| CEP170 | NOX4 | 0.755938 | 3 | 0 | 3 |
| CEP170 | LAMB1 | 0.754298 | 3 | 0 | 3 |
| CEP170 | PXDN | 0.748989 | 3 | 0 | 3 |
| CEP170 | MEX3B | 0.748726 | 3 | 0 | 3 |
| CEP170 | CHN1 | 0.746551 | 3 | 0 | 3 |
| CEP170 | LPCAT1 | 0.746295 | 3 | 0 | 3 |
| CEP170 | LRP12 | 0.744871 | 3 | 0 | 3 |
| CEP170 | MMD | 0.742462 | 3 | 0 | 3 |
| CEP170 | HMGN4 | 0.742093 | 3 | 0 | 3 |
| CEP170 | RNF2 | 0.741951 | 3 | 0 | 3 |
| CEP170 | SUPT16H | 0.735536 | 3 | 0 | 3 |
| CEP170 | HEATR1 | 0.734678 | 3 | 0 | 3 |
| CEP170 | RAB34 | 0.734155 | 3 | 0 | 3 |
| CEP170 | RCN1 | 0.731509 | 3 | 0 | 3 |
For details and further investigation, click here