| Full name: fatty acid desaturase 1 | Alias Symbol: D5D|FADSD5|TU12|FADS6 | ||
| Type: protein-coding gene | Cytoband: 11q12.2 | ||
| Entrez ID: 3992 | HGNC ID: HGNC:3574 | Ensembl Gene: ENSG00000149485 | OMIM ID: 606148 |
| Drug and gene relationship at DGIdb | |||
Expression of FADS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | FADS1 | 3992 | 46168 | 1.7977 | 0.0000 | |
| GSE53624 | FADS1 | 3992 | 46168 | 2.3224 | 0.0000 | |
| GSE97050 | FADS1 | 3992 | A_24_P192994 | 0.3144 | 0.5391 | |
| SRP007169 | FADS1 | 3992 | RNAseq | 4.6281 | 0.0000 | |
| SRP008496 | FADS1 | 3992 | RNAseq | 4.5745 | 0.0000 | |
| SRP064894 | FADS1 | 3992 | RNAseq | 1.8847 | 0.0000 | |
| SRP133303 | FADS1 | 3992 | RNAseq | 1.6061 | 0.0000 | |
| SRP159526 | FADS1 | 3992 | RNAseq | 2.9857 | 0.0000 | |
| SRP193095 | FADS1 | 3992 | RNAseq | 2.8230 | 0.0000 | |
| SRP219564 | FADS1 | 3992 | RNAseq | 1.7038 | 0.0010 | |
| TCGA | FADS1 | 3992 | RNAseq | 0.3993 | 0.0042 |
Upregulated datasets: 9; Downregulated datasets: 0.
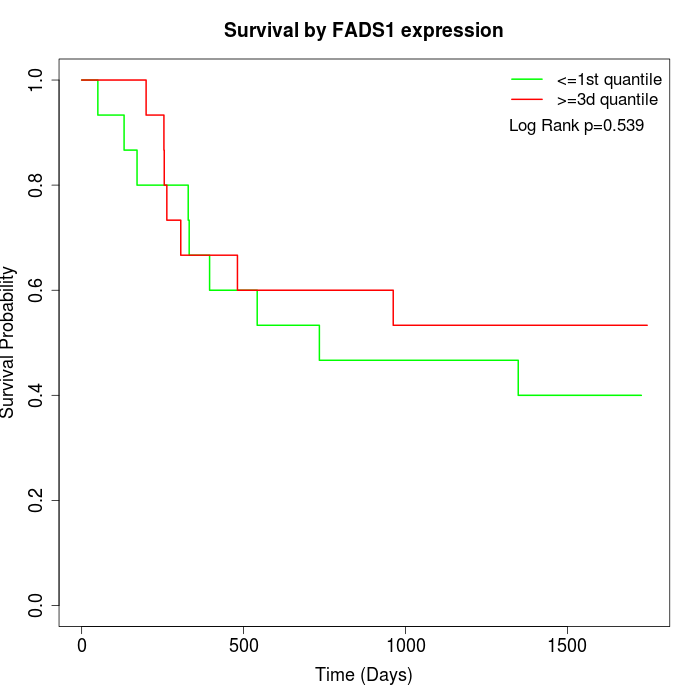
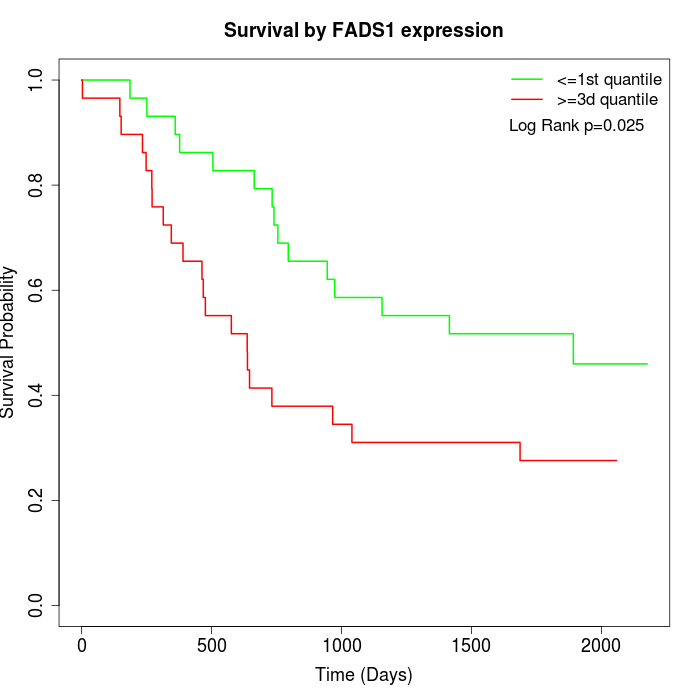
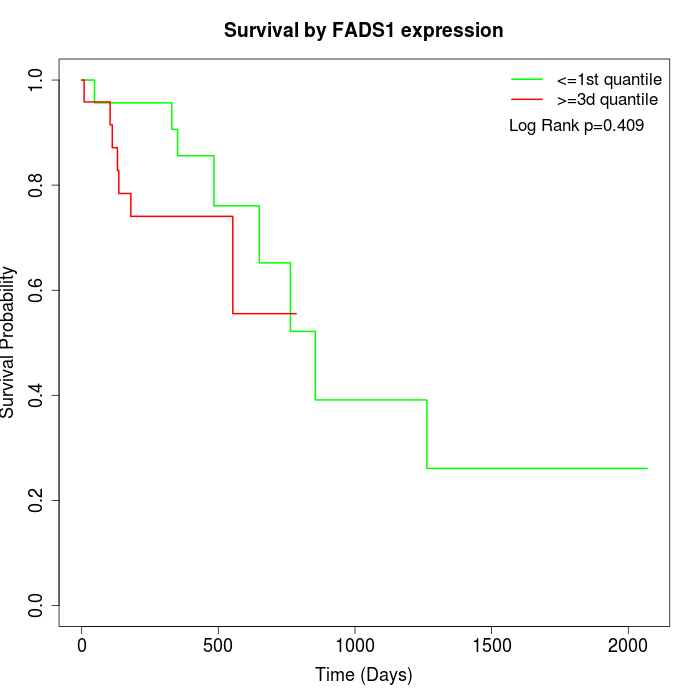
Survival by FADS1 expression:
Note: Click image to view full size file.
Copy number change of FADS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FADS1 | 3992 | 5 | 5 | 20 | |
| GSE20123 | FADS1 | 3992 | 5 | 5 | 20 | |
| GSE43470 | FADS1 | 3992 | 2 | 6 | 35 | |
| GSE46452 | FADS1 | 3992 | 8 | 4 | 47 | |
| GSE47630 | FADS1 | 3992 | 3 | 7 | 30 | |
| GSE54993 | FADS1 | 3992 | 3 | 0 | 67 | |
| GSE54994 | FADS1 | 3992 | 4 | 8 | 41 | |
| GSE60625 | FADS1 | 3992 | 0 | 3 | 8 | |
| GSE74703 | FADS1 | 3992 | 2 | 3 | 31 | |
| GSE74704 | FADS1 | 3992 | 4 | 3 | 13 | |
| TCGA | FADS1 | 3992 | 14 | 10 | 72 |
Total number of gains: 50; Total number of losses: 54; Total Number of normals: 384.
Somatic mutations of FADS1:
Generating mutation plots.
Highly correlated genes for FADS1:
Showing top 20/794 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FADS1 | LPCAT1 | 0.843774 | 3 | 0 | 3 |
| FADS1 | WDR54 | 0.823676 | 3 | 0 | 3 |
| FADS1 | RAB34 | 0.823053 | 3 | 0 | 3 |
| FADS1 | LAPTM4B | 0.820822 | 3 | 0 | 3 |
| FADS1 | PLOD1 | 0.820178 | 3 | 0 | 3 |
| FADS1 | KIF3C | 0.815294 | 3 | 0 | 3 |
| FADS1 | MMD | 0.809429 | 3 | 0 | 3 |
| FADS1 | FUS | 0.80813 | 3 | 0 | 3 |
| FADS1 | PXDN | 0.805662 | 3 | 0 | 3 |
| FADS1 | TP53I13 | 0.805318 | 3 | 0 | 3 |
| FADS1 | FARP1 | 0.803972 | 3 | 0 | 3 |
| FADS1 | CHN1 | 0.802289 | 3 | 0 | 3 |
| FADS1 | ATP6V1C1 | 0.802274 | 3 | 0 | 3 |
| FADS1 | GNA12 | 0.801602 | 3 | 0 | 3 |
| FADS1 | RAI14 | 0.800891 | 3 | 0 | 3 |
| FADS1 | TNFAIP6 | 0.799719 | 3 | 0 | 3 |
| FADS1 | RCN1 | 0.798776 | 3 | 0 | 3 |
| FADS1 | ENAH | 0.796471 | 3 | 0 | 3 |
| FADS1 | PFDN2 | 0.793618 | 3 | 0 | 3 |
| FADS1 | YWHAH | 0.7934 | 3 | 0 | 3 |
For details and further investigation, click here