| Full name: G protein subunit gamma 10 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 9q31.3 | ||
| Entrez ID: 2790 | HGNC ID: HGNC:4402 | Ensembl Gene: ENSG00000242616 | OMIM ID: 604389 |
| Drug and gene relationship at DGIdb | |||
GNG10 involved pathways:
Expression of GNG10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | GNG10 | 2790 | 89382 | 0.8356 | 0.0000 | |
| GSE53624 | GNG10 | 2790 | 89382 | 0.7877 | 0.0000 | |
| GSE97050 | GNG10 | 2790 | A_23_P112260 | 0.7730 | 0.1033 | |
| SRP007169 | GNG10 | 2790 | RNAseq | 0.9701 | 0.0195 | |
| SRP008496 | GNG10 | 2790 | RNAseq | 1.0515 | 0.0000 | |
| SRP064894 | GNG10 | 2790 | RNAseq | 0.9285 | 0.0000 | |
| SRP133303 | GNG10 | 2790 | RNAseq | 1.1108 | 0.0000 | |
| SRP159526 | GNG10 | 2790 | RNAseq | 0.5331 | 0.0061 | |
| SRP193095 | GNG10 | 2790 | RNAseq | 0.2105 | 0.1518 | |
| SRP219564 | GNG10 | 2790 | RNAseq | 0.4539 | 0.3930 | |
| TCGA | GNG10 | 2790 | RNAseq | -0.0094 | 0.8874 |
Upregulated datasets: 2; Downregulated datasets: 0.
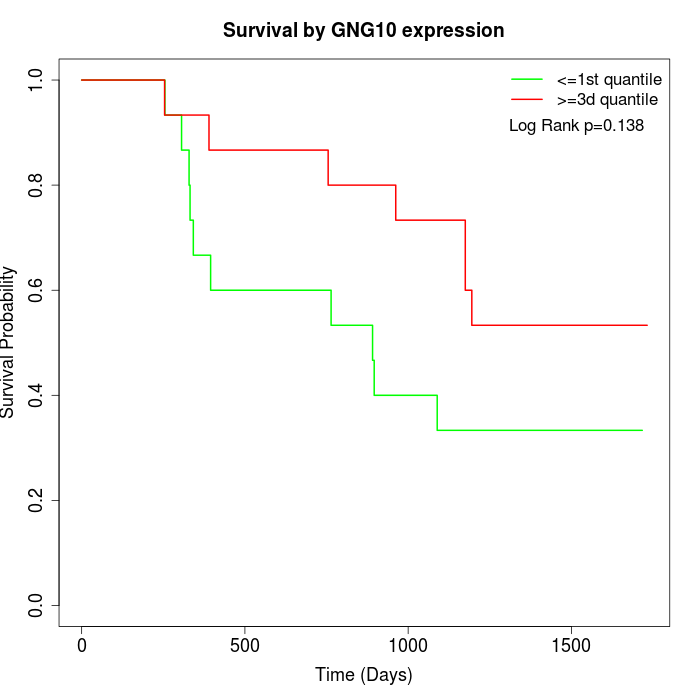
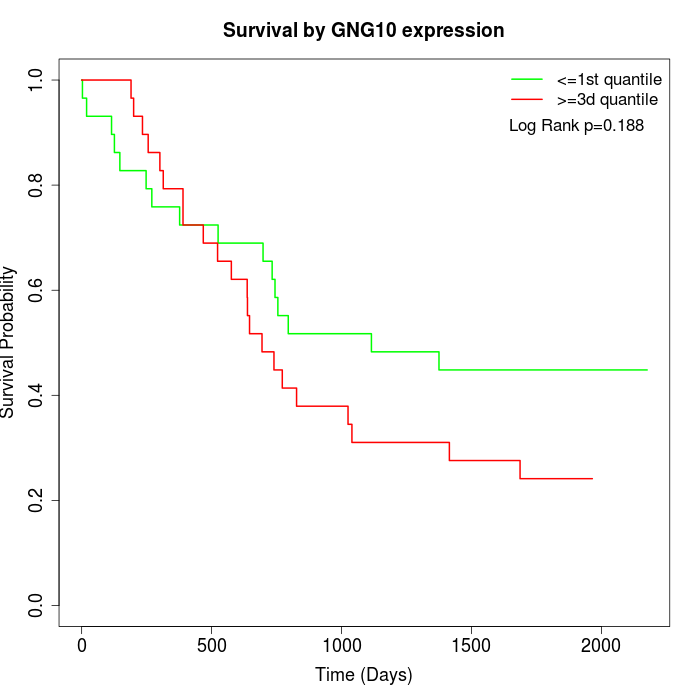
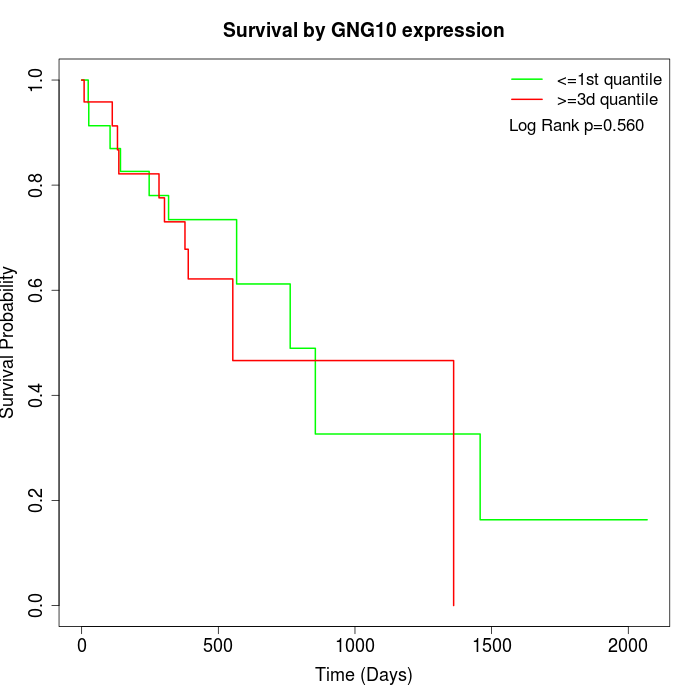
Survival by GNG10 expression:
Note: Click image to view full size file.
Copy number change of GNG10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GNG10 | 2790 | 5 | 8 | 17 | |
| GSE20123 | GNG10 | 2790 | 5 | 7 | 18 | |
| GSE43470 | GNG10 | 2790 | 4 | 3 | 36 | |
| GSE46452 | GNG10 | 2790 | 6 | 13 | 40 | |
| GSE47630 | GNG10 | 2790 | 3 | 16 | 21 | |
| GSE54993 | GNG10 | 2790 | 3 | 3 | 64 | |
| GSE54994 | GNG10 | 2790 | 9 | 10 | 34 | |
| GSE60625 | GNG10 | 2790 | 0 | 0 | 11 | |
| GSE74703 | GNG10 | 2790 | 4 | 3 | 29 | |
| GSE74704 | GNG10 | 2790 | 2 | 5 | 13 | |
| TCGA | GNG10 | 2790 | 26 | 21 | 49 |
Total number of gains: 67; Total number of losses: 89; Total Number of normals: 332.
Somatic mutations of GNG10:
Generating mutation plots.
Highly correlated genes for GNG10:
Showing top 20/395 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GNG10 | CALU | 0.812105 | 3 | 0 | 3 |
| GNG10 | LPCAT1 | 0.811749 | 3 | 0 | 3 |
| GNG10 | SRSF10 | 0.794321 | 3 | 0 | 3 |
| GNG10 | MSN | 0.792431 | 3 | 0 | 3 |
| GNG10 | ACLY | 0.790242 | 3 | 0 | 3 |
| GNG10 | ADAR | 0.784759 | 3 | 0 | 3 |
| GNG10 | GNA13 | 0.781951 | 3 | 0 | 3 |
| GNG10 | PXDN | 0.778051 | 3 | 0 | 3 |
| GNG10 | GNA12 | 0.774508 | 3 | 0 | 3 |
| GNG10 | TRAM2 | 0.773934 | 3 | 0 | 3 |
| GNG10 | P4HA1 | 0.772353 | 3 | 0 | 3 |
| GNG10 | CBLB | 0.769034 | 3 | 0 | 3 |
| GNG10 | FZD2 | 0.768779 | 3 | 0 | 3 |
| GNG10 | STK3 | 0.767093 | 3 | 0 | 3 |
| GNG10 | HEATR6 | 0.766834 | 3 | 0 | 3 |
| GNG10 | MORC2 | 0.764918 | 3 | 0 | 3 |
| GNG10 | ASAP1 | 0.763686 | 3 | 0 | 3 |
| GNG10 | PPT1 | 0.763243 | 3 | 0 | 3 |
| GNG10 | LAMB1 | 0.762158 | 3 | 0 | 3 |
| GNG10 | RNF2 | 0.760689 | 3 | 0 | 3 |
For details and further investigation, click here