| Full name: laminin subunit beta 2 | Alias Symbol: NPHS5 | ||
| Type: protein-coding gene | Cytoband: 3p21.31 | ||
| Entrez ID: 3913 | HGNC ID: HGNC:6487 | Ensembl Gene: ENSG00000172037 | OMIM ID: 150325 |
| Drug and gene relationship at DGIdb | |||
LAMB2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04510 | Focal adhesion | |
| hsa05200 | Pathways in cancer | |
| hsa05222 | Small cell lung cancer |
Expression of LAMB2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LAMB2 | 3913 | 216264_s_at | -0.7157 | 0.2453 | |
| GSE20347 | LAMB2 | 3913 | 216264_s_at | -0.4696 | 0.0055 | |
| GSE23400 | LAMB2 | 3913 | 216264_s_at | -0.4464 | 0.0014 | |
| GSE26886 | LAMB2 | 3913 | 216264_s_at | 0.8522 | 0.0003 | |
| GSE29001 | LAMB2 | 3913 | 216264_s_at | -0.1160 | 0.8166 | |
| GSE38129 | LAMB2 | 3913 | 216264_s_at | -0.8168 | 0.0091 | |
| GSE45670 | LAMB2 | 3913 | 216264_s_at | -0.7113 | 0.0001 | |
| GSE53622 | LAMB2 | 3913 | 32367 | -0.5403 | 0.0000 | |
| GSE53624 | LAMB2 | 3913 | 32367 | -0.2645 | 0.0008 | |
| GSE63941 | LAMB2 | 3913 | 216264_s_at | -1.4729 | 0.0196 | |
| GSE77861 | LAMB2 | 3913 | 216264_s_at | -0.1369 | 0.6317 | |
| GSE97050 | LAMB2 | 3913 | A_33_P3223780 | -0.6575 | 0.2263 | |
| SRP007169 | LAMB2 | 3913 | RNAseq | 1.1583 | 0.0114 | |
| SRP008496 | LAMB2 | 3913 | RNAseq | 1.3040 | 0.0000 | |
| SRP064894 | LAMB2 | 3913 | RNAseq | 0.4420 | 0.1041 | |
| SRP133303 | LAMB2 | 3913 | RNAseq | -0.3795 | 0.2248 | |
| SRP159526 | LAMB2 | 3913 | RNAseq | -0.4286 | 0.1036 | |
| SRP193095 | LAMB2 | 3913 | RNAseq | 0.2050 | 0.2046 | |
| SRP219564 | LAMB2 | 3913 | RNAseq | -0.2490 | 0.6830 | |
| TCGA | LAMB2 | 3913 | RNAseq | -0.4298 | 0.0000 |
Upregulated datasets: 2; Downregulated datasets: 1.
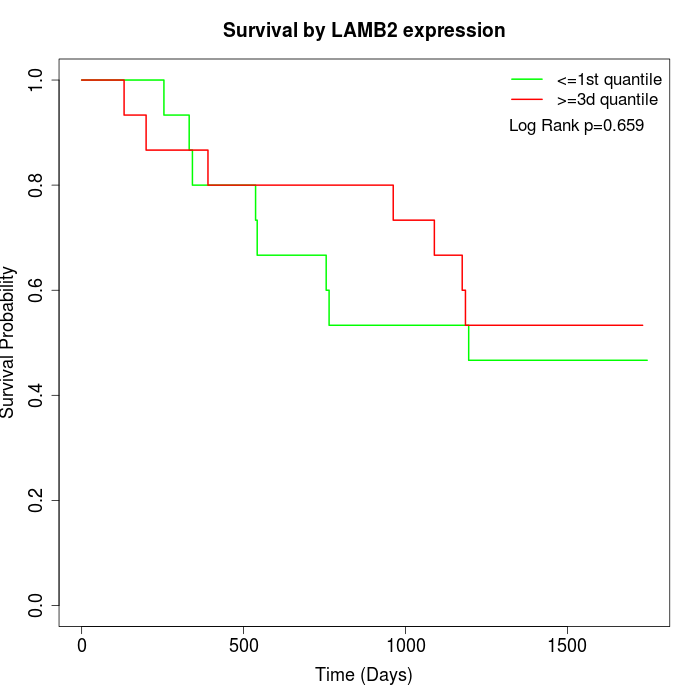
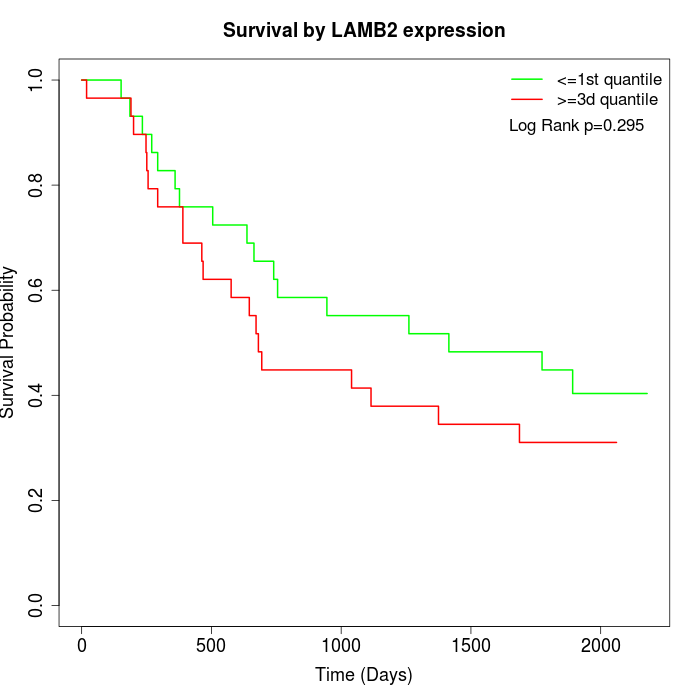
Survival by LAMB2 expression:
Note: Click image to view full size file.
Copy number change of LAMB2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LAMB2 | 3913 | 0 | 18 | 12 | |
| GSE20123 | LAMB2 | 3913 | 0 | 19 | 11 | |
| GSE43470 | LAMB2 | 3913 | 0 | 18 | 25 | |
| GSE46452 | LAMB2 | 3913 | 2 | 16 | 41 | |
| GSE47630 | LAMB2 | 3913 | 1 | 24 | 15 | |
| GSE54993 | LAMB2 | 3913 | 6 | 2 | 62 | |
| GSE54994 | LAMB2 | 3913 | 0 | 36 | 17 | |
| GSE60625 | LAMB2 | 3913 | 5 | 0 | 6 | |
| GSE74703 | LAMB2 | 3913 | 0 | 14 | 22 | |
| GSE74704 | LAMB2 | 3913 | 0 | 12 | 8 | |
| TCGA | LAMB2 | 3913 | 0 | 78 | 18 |
Total number of gains: 14; Total number of losses: 237; Total Number of normals: 237.
Somatic mutations of LAMB2:
Generating mutation plots.
Highly correlated genes for LAMB2:
Showing top 20/854 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LAMB2 | LAMA2 | 0.745898 | 8 | 0 | 8 |
| LAMB2 | FBXL7 | 0.732382 | 9 | 0 | 8 |
| LAMB2 | C1QTNF2 | 0.723952 | 5 | 0 | 5 |
| LAMB2 | FOXF2 | 0.722977 | 8 | 0 | 8 |
| LAMB2 | PGR | 0.719856 | 5 | 0 | 5 |
| LAMB2 | ADAMTSL1 | 0.717 | 7 | 0 | 6 |
| LAMB2 | TCEAL3 | 0.716762 | 3 | 0 | 3 |
| LAMB2 | CYS1 | 0.716587 | 6 | 0 | 5 |
| LAMB2 | ILK | 0.716372 | 9 | 0 | 9 |
| LAMB2 | NKX6-1 | 0.714618 | 6 | 0 | 6 |
| LAMB2 | ARHGEF25 | 0.711876 | 5 | 0 | 5 |
| LAMB2 | SORCS1 | 0.711735 | 4 | 0 | 4 |
| LAMB2 | NEGR1 | 0.710116 | 6 | 0 | 6 |
| LAMB2 | RAI2 | 0.709333 | 9 | 0 | 8 |
| LAMB2 | TLN1 | 0.709117 | 6 | 0 | 6 |
| LAMB2 | DPT | 0.707084 | 9 | 0 | 7 |
| LAMB2 | UBXN10 | 0.707007 | 3 | 0 | 3 |
| LAMB2 | TCEAL1 | 0.705758 | 7 | 0 | 7 |
| LAMB2 | PARVA | 0.703963 | 10 | 0 | 9 |
| LAMB2 | RBM24 | 0.703894 | 5 | 0 | 4 |
For details and further investigation, click here