| Full name: neuronal growth regulator 1 | Alias Symbol: KILON|MGC46680|Ntra|IGLON4 | ||
| Type: protein-coding gene | Cytoband: 1p31.1 | ||
| Entrez ID: 257194 | HGNC ID: HGNC:17302 | Ensembl Gene: ENSG00000172260 | OMIM ID: 613173 |
| Drug and gene relationship at DGIdb | |||
Expression of NEGR1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NEGR1 | 257194 | 229461_x_at | -2.8925 | 0.0756 | |
| GSE26886 | NEGR1 | 257194 | 243357_at | -0.0623 | 0.7745 | |
| GSE45670 | NEGR1 | 257194 | 243357_at | -2.2830 | 0.0000 | |
| GSE53622 | NEGR1 | 257194 | 67680 | -1.8150 | 0.0000 | |
| GSE53624 | NEGR1 | 257194 | 67680 | -1.7775 | 0.0000 | |
| GSE63941 | NEGR1 | 257194 | 243357_at | -3.5925 | 0.0000 | |
| GSE77861 | NEGR1 | 257194 | 243357_at | -0.0120 | 0.8945 | |
| GSE97050 | NEGR1 | 257194 | A_33_P3341970 | -1.1351 | 0.1453 | |
| SRP007169 | NEGR1 | 257194 | RNAseq | 0.9582 | 0.0861 | |
| SRP008496 | NEGR1 | 257194 | RNAseq | 1.9157 | 0.0002 | |
| SRP064894 | NEGR1 | 257194 | RNAseq | -0.5196 | 0.1321 | |
| SRP133303 | NEGR1 | 257194 | RNAseq | -0.1608 | 0.3733 | |
| SRP159526 | NEGR1 | 257194 | RNAseq | -0.8491 | 0.0004 | |
| SRP193095 | NEGR1 | 257194 | RNAseq | 0.3903 | 0.0056 | |
| SRP219564 | NEGR1 | 257194 | RNAseq | -0.7486 | 0.3665 | |
| TCGA | NEGR1 | 257194 | RNAseq | -1.2780 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 5.
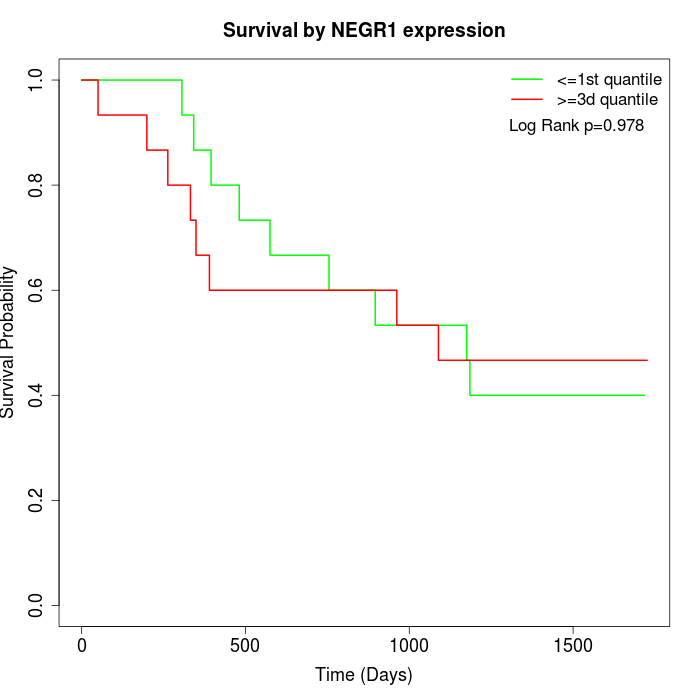
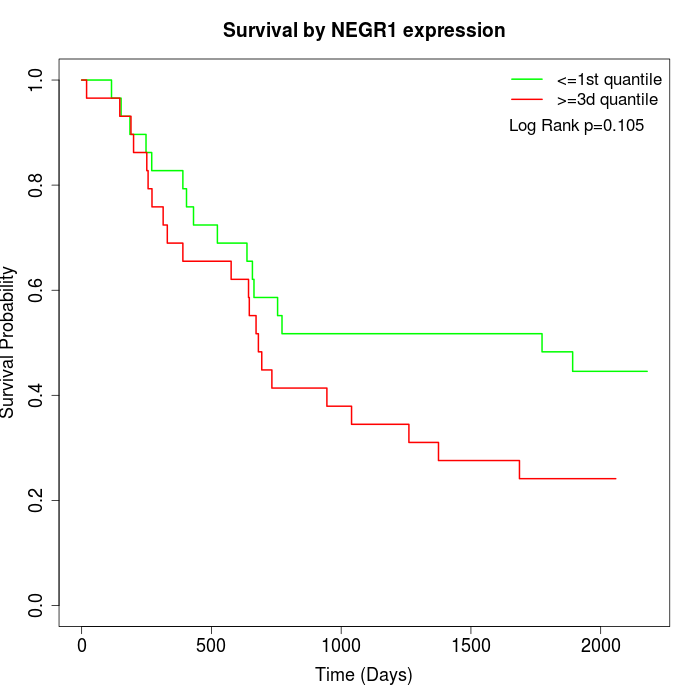
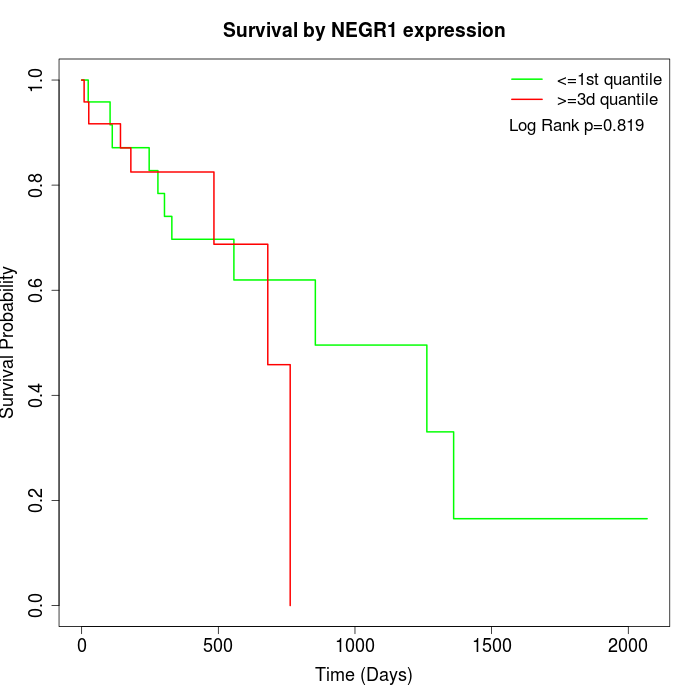
Survival by NEGR1 expression:
Note: Click image to view full size file.
Copy number change of NEGR1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NEGR1 | 257194 | 1 | 7 | 22 | |
| GSE20123 | NEGR1 | 257194 | 1 | 7 | 22 | |
| GSE43470 | NEGR1 | 257194 | 4 | 2 | 37 | |
| GSE46452 | NEGR1 | 257194 | 0 | 1 | 58 | |
| GSE47630 | NEGR1 | 257194 | 9 | 7 | 24 | |
| GSE54993 | NEGR1 | 257194 | 0 | 1 | 69 | |
| GSE54994 | NEGR1 | 257194 | 6 | 5 | 42 | |
| GSE60625 | NEGR1 | 257194 | 0 | 0 | 11 | |
| GSE74703 | NEGR1 | 257194 | 3 | 2 | 31 | |
| GSE74704 | NEGR1 | 257194 | 1 | 4 | 15 | |
| TCGA | NEGR1 | 257194 | 8 | 24 | 64 |
Total number of gains: 33; Total number of losses: 60; Total Number of normals: 395.
Somatic mutations of NEGR1:
Generating mutation plots.
Highly correlated genes for NEGR1:
Showing top 20/1364 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NEGR1 | PGM5 | 0.909552 | 5 | 0 | 5 |
| NEGR1 | MRVI1 | 0.888629 | 6 | 0 | 6 |
| NEGR1 | NEXN | 0.887324 | 6 | 0 | 6 |
| NEGR1 | PGM5-AS1 | 0.873506 | 3 | 0 | 3 |
| NEGR1 | DPT | 0.864262 | 7 | 0 | 7 |
| NEGR1 | LINC01082 | 0.861107 | 5 | 0 | 5 |
| NEGR1 | COX7A1 | 0.8595 | 6 | 0 | 6 |
| NEGR1 | FXYD1 | 0.858537 | 4 | 0 | 4 |
| NEGR1 | FENDRR | 0.857511 | 5 | 0 | 5 |
| NEGR1 | MAGI2-AS3 | 0.856181 | 5 | 0 | 5 |
| NEGR1 | OGN | 0.855864 | 6 | 0 | 6 |
| NEGR1 | MYOM1 | 0.855093 | 5 | 0 | 5 |
| NEGR1 | AGTR1 | 0.854883 | 6 | 0 | 6 |
| NEGR1 | SMIM10 | 0.848356 | 5 | 0 | 5 |
| NEGR1 | CACNB2 | 0.847334 | 5 | 0 | 5 |
| NEGR1 | CYP21A2 | 0.846843 | 3 | 0 | 3 |
| NEGR1 | CCDC69 | 0.846172 | 6 | 0 | 6 |
| NEGR1 | ABCA6 | 0.84568 | 6 | 0 | 6 |
| NEGR1 | DIXDC1 | 0.840939 | 6 | 0 | 6 |
| NEGR1 | LRRN4CL | 0.840329 | 6 | 0 | 6 |
For details and further investigation, click here