| Full name: lipocalin 10 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 9q34.3 | ||
| Entrez ID: 414332 | HGNC ID: HGNC:20892 | Ensembl Gene: ENSG00000187922 | OMIM ID: 612904 |
| Drug and gene relationship at DGIdb | |||
Expression of LCN10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LCN10 | 414332 | 238071_at | -0.0184 | 0.9718 | |
| GSE26886 | LCN10 | 414332 | 238071_at | 0.0280 | 0.8749 | |
| GSE45670 | LCN10 | 414332 | 238071_at | -0.0200 | 0.8437 | |
| GSE63941 | LCN10 | 414332 | 238071_at | 0.1197 | 0.5488 | |
| GSE77861 | LCN10 | 414332 | 238071_at | 0.0020 | 0.9876 | |
| GSE97050 | LCN10 | 414332 | A_33_P3263955 | 0.3973 | 0.2682 | |
| SRP133303 | LCN10 | 414332 | RNAseq | -2.3960 | 0.0000 | |
| SRP159526 | LCN10 | 414332 | RNAseq | -2.1366 | 0.0000 | |
| SRP219564 | LCN10 | 414332 | RNAseq | -1.2723 | 0.1549 | |
| TCGA | LCN10 | 414332 | RNAseq | -2.5458 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 3.
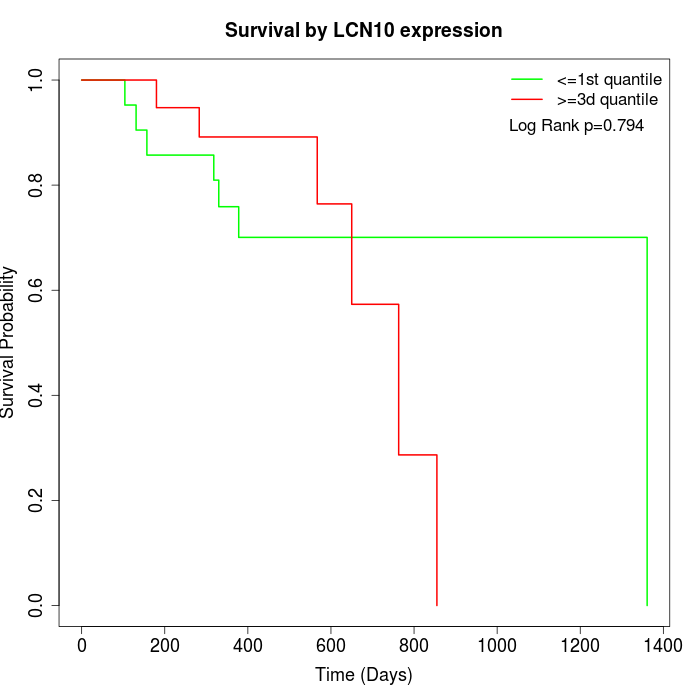
Survival by LCN10 expression:
Note: Click image to view full size file.
Copy number change of LCN10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LCN10 | 414332 | 5 | 7 | 18 | |
| GSE20123 | LCN10 | 414332 | 5 | 7 | 18 | |
| GSE43470 | LCN10 | 414332 | 3 | 7 | 33 | |
| GSE46452 | LCN10 | 414332 | 6 | 13 | 40 | |
| GSE47630 | LCN10 | 414332 | 6 | 15 | 19 | |
| GSE54993 | LCN10 | 414332 | 3 | 3 | 64 | |
| GSE54994 | LCN10 | 414332 | 12 | 8 | 33 | |
| GSE60625 | LCN10 | 414332 | 0 | 0 | 11 | |
| GSE74703 | LCN10 | 414332 | 3 | 5 | 28 | |
| GSE74704 | LCN10 | 414332 | 3 | 5 | 12 | |
| TCGA | LCN10 | 414332 | 29 | 23 | 44 |
Total number of gains: 75; Total number of losses: 93; Total Number of normals: 320.
Somatic mutations of LCN10:
Generating mutation plots.
Highly correlated genes for LCN10:
Showing top 20/92 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LCN10 | SMPDL3B | 0.724532 | 3 | 0 | 3 |
| LCN10 | ZNF747 | 0.708337 | 3 | 0 | 3 |
| LCN10 | NDUFAF3 | 0.684121 | 3 | 0 | 3 |
| LCN10 | GGT5 | 0.670008 | 4 | 0 | 3 |
| LCN10 | GBX1 | 0.667782 | 3 | 0 | 3 |
| LCN10 | FBXW5 | 0.666455 | 3 | 0 | 3 |
| LCN10 | LINC00642 | 0.661275 | 3 | 0 | 3 |
| LCN10 | CSF1 | 0.657743 | 4 | 0 | 3 |
| LCN10 | ENTPD2 | 0.656856 | 3 | 0 | 3 |
| LCN10 | FAM167B | 0.656735 | 3 | 0 | 3 |
| LCN10 | ADPRH | 0.653978 | 3 | 0 | 3 |
| LCN10 | ARHGEF15 | 0.652773 | 4 | 0 | 4 |
| LCN10 | ZNF345 | 0.649185 | 3 | 0 | 3 |
| LCN10 | EXOC3L2 | 0.648632 | 4 | 0 | 4 |
| LCN10 | CNTNAP1 | 0.648006 | 3 | 0 | 3 |
| LCN10 | DENND5B | 0.643208 | 3 | 0 | 3 |
| LCN10 | C22orf31 | 0.64314 | 3 | 0 | 3 |
| LCN10 | PARM1 | 0.642855 | 3 | 0 | 3 |
| LCN10 | RNASEH2C | 0.638906 | 3 | 0 | 3 |
| LCN10 | INSRR | 0.637853 | 3 | 0 | 3 |
For details and further investigation, click here