| Full name: gastrulation brain homeobox 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 7q36.1 | ||
| Entrez ID: 2636 | HGNC ID: HGNC:4185 | Ensembl Gene: ENSG00000164900 | OMIM ID: 603354 |
| Drug and gene relationship at DGIdb | |||
Expression of GBX1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GBX1 | 2636 | 217011_at | 0.0148 | 0.9632 | |
| GSE20347 | GBX1 | 2636 | 217011_at | -0.0819 | 0.2570 | |
| GSE23400 | GBX1 | 2636 | 217011_at | -0.0886 | 0.0085 | |
| GSE26886 | GBX1 | 2636 | 217011_at | 0.1450 | 0.2151 | |
| GSE29001 | GBX1 | 2636 | 217011_at | -0.2169 | 0.2074 | |
| GSE38129 | GBX1 | 2636 | 217011_at | -0.1253 | 0.0673 | |
| GSE45670 | GBX1 | 2636 | 217011_at | 0.0181 | 0.8475 | |
| GSE53622 | GBX1 | 2636 | 35860 | -0.0493 | 0.5910 | |
| GSE53624 | GBX1 | 2636 | 35860 | -0.0896 | 0.0840 | |
| GSE63941 | GBX1 | 2636 | 217011_at | 0.2844 | 0.0673 | |
| GSE77861 | GBX1 | 2636 | 217011_at | 0.0079 | 0.9571 | |
| TCGA | GBX1 | 2636 | RNAseq | 0.8023 | 0.6050 |
Upregulated datasets: 0; Downregulated datasets: 0.
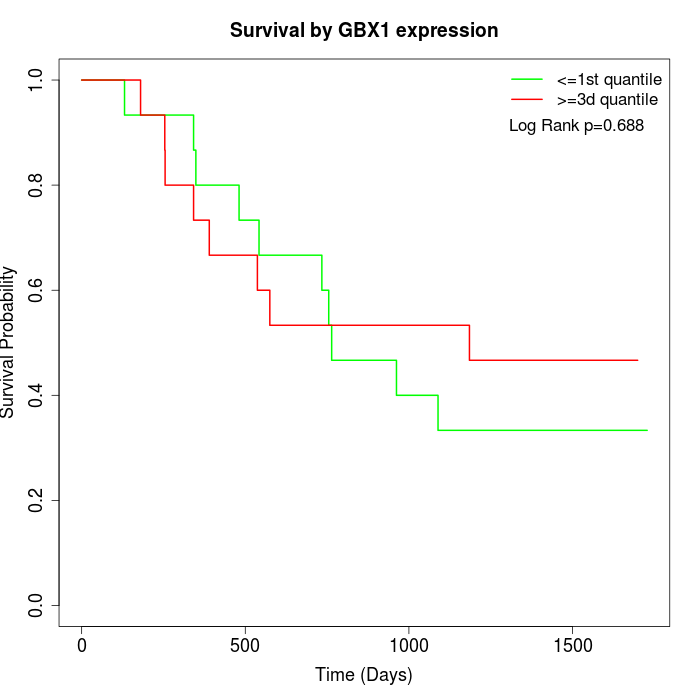
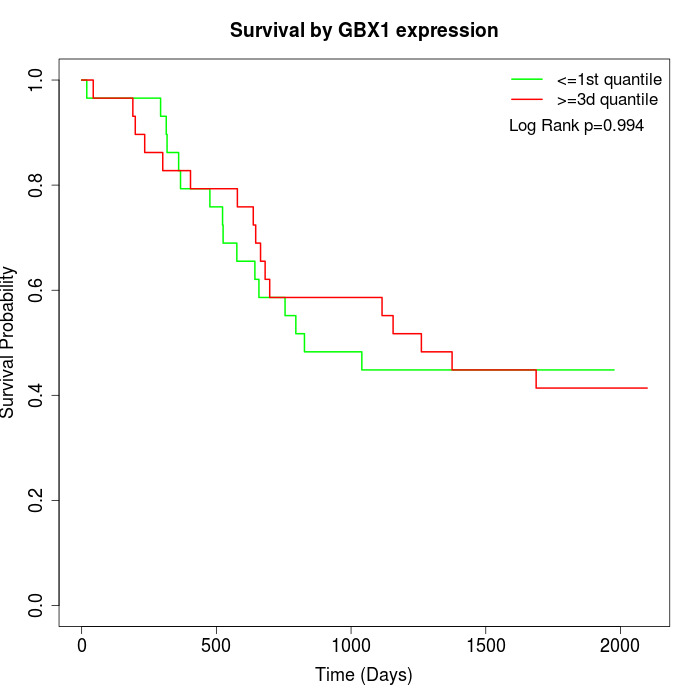
Survival by GBX1 expression:
Note: Click image to view full size file.
Copy number change of GBX1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GBX1 | 2636 | 2 | 4 | 24 | |
| GSE20123 | GBX1 | 2636 | 2 | 4 | 24 | |
| GSE43470 | GBX1 | 2636 | 2 | 5 | 36 | |
| GSE46452 | GBX1 | 2636 | 7 | 2 | 50 | |
| GSE47630 | GBX1 | 2636 | 5 | 9 | 26 | |
| GSE54993 | GBX1 | 2636 | 3 | 3 | 64 | |
| GSE54994 | GBX1 | 2636 | 5 | 8 | 40 | |
| GSE60625 | GBX1 | 2636 | 0 | 0 | 11 | |
| GSE74703 | GBX1 | 2636 | 2 | 4 | 30 | |
| GSE74704 | GBX1 | 2636 | 1 | 4 | 15 | |
| TCGA | GBX1 | 2636 | 28 | 25 | 43 |
Total number of gains: 57; Total number of losses: 68; Total Number of normals: 363.
Somatic mutations of GBX1:
Generating mutation plots.
Highly correlated genes for GBX1:
Showing top 20/1145 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GBX1 | IL18BP | 0.714937 | 4 | 0 | 4 |
| GBX1 | KIR2DS3 | 0.703727 | 5 | 0 | 5 |
| GBX1 | BOLA2 | 0.691842 | 3 | 0 | 3 |
| GBX1 | PI16 | 0.690839 | 3 | 0 | 3 |
| GBX1 | KLF15 | 0.689 | 4 | 0 | 4 |
| GBX1 | ADM2 | 0.682517 | 7 | 0 | 6 |
| GBX1 | AAAS | 0.67969 | 4 | 0 | 4 |
| GBX1 | PF4 | 0.677866 | 3 | 0 | 3 |
| GBX1 | GPR17 | 0.673719 | 4 | 0 | 4 |
| GBX1 | NKAIN4 | 0.672176 | 3 | 0 | 3 |
| GBX1 | AMHR2 | 0.671787 | 7 | 0 | 6 |
| GBX1 | EDA2R | 0.670545 | 5 | 0 | 5 |
| GBX1 | BMP15 | 0.670542 | 6 | 0 | 6 |
| GBX1 | PHF7 | 0.667888 | 4 | 0 | 4 |
| GBX1 | LCN10 | 0.667782 | 3 | 0 | 3 |
| GBX1 | ASIC4 | 0.664044 | 7 | 0 | 7 |
| GBX1 | MYO15B | 0.663102 | 3 | 0 | 3 |
| GBX1 | KLF16 | 0.662594 | 4 | 0 | 4 |
| GBX1 | PAX7 | 0.660949 | 4 | 0 | 4 |
| GBX1 | ANGPT4 | 0.660168 | 6 | 0 | 6 |
For details and further investigation, click here