| Full name: LIF receptor subunit alpha | Alias Symbol: CD118 | ||
| Type: protein-coding gene | Cytoband: 5p13.1 | ||
| Entrez ID: 3977 | HGNC ID: HGNC:6597 | Ensembl Gene: ENSG00000113594 | OMIM ID: 151443 |
| Drug and gene relationship at DGIdb | |||
LIFR involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04550 | Signaling pathways regulating pluripotency of stem cells | |
| hsa04630 | Jak-STAT signaling pathway |
Expression of LIFR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LIFR | 3977 | 225575_at | -2.2503 | 0.0475 | |
| GSE20347 | LIFR | 3977 | 205876_at | 0.0917 | 0.4573 | |
| GSE23400 | LIFR | 3977 | 205876_at | -0.0157 | 0.8133 | |
| GSE26886 | LIFR | 3977 | 225575_at | 0.9006 | 0.0525 | |
| GSE29001 | LIFR | 3977 | 205876_at | 0.2679 | 0.4476 | |
| GSE38129 | LIFR | 3977 | 205876_at | -0.1849 | 0.4173 | |
| GSE45670 | LIFR | 3977 | 225575_at | -2.7166 | 0.0000 | |
| GSE53622 | LIFR | 3977 | 12991 | -1.8641 | 0.0000 | |
| GSE53624 | LIFR | 3977 | 12991 | -1.7142 | 0.0000 | |
| GSE63941 | LIFR | 3977 | 225575_at | -0.4349 | 0.6165 | |
| GSE77861 | LIFR | 3977 | 227771_at | 0.0710 | 0.6858 | |
| GSE97050 | LIFR | 3977 | A_24_P397386 | -0.9193 | 0.0958 | |
| SRP007169 | LIFR | 3977 | RNAseq | 0.6983 | 0.3325 | |
| SRP008496 | LIFR | 3977 | RNAseq | 0.8346 | 0.0702 | |
| SRP064894 | LIFR | 3977 | RNAseq | -1.5438 | 0.0003 | |
| SRP133303 | LIFR | 3977 | RNAseq | -1.7777 | 0.0000 | |
| SRP193095 | LIFR | 3977 | RNAseq | -0.6014 | 0.0823 | |
| SRP219564 | LIFR | 3977 | RNAseq | -1.5258 | 0.0078 | |
| TCGA | LIFR | 3977 | RNAseq | -1.0621 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 8.
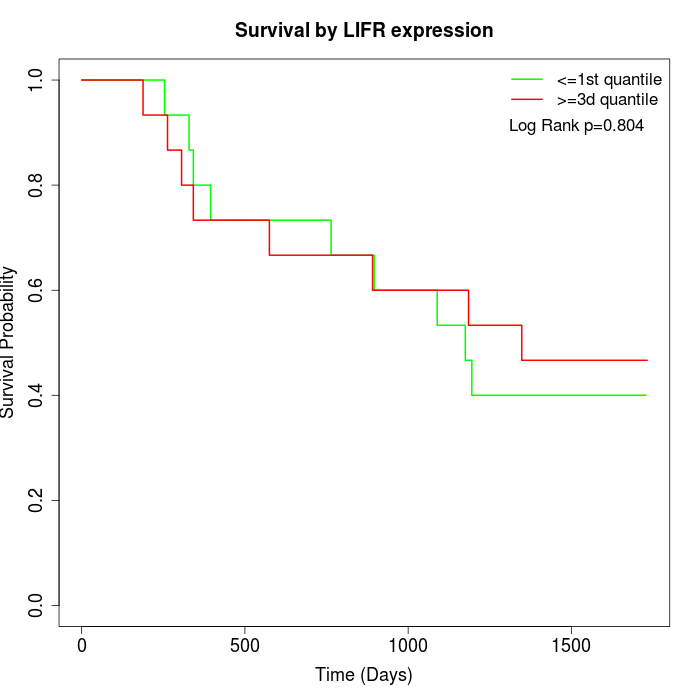
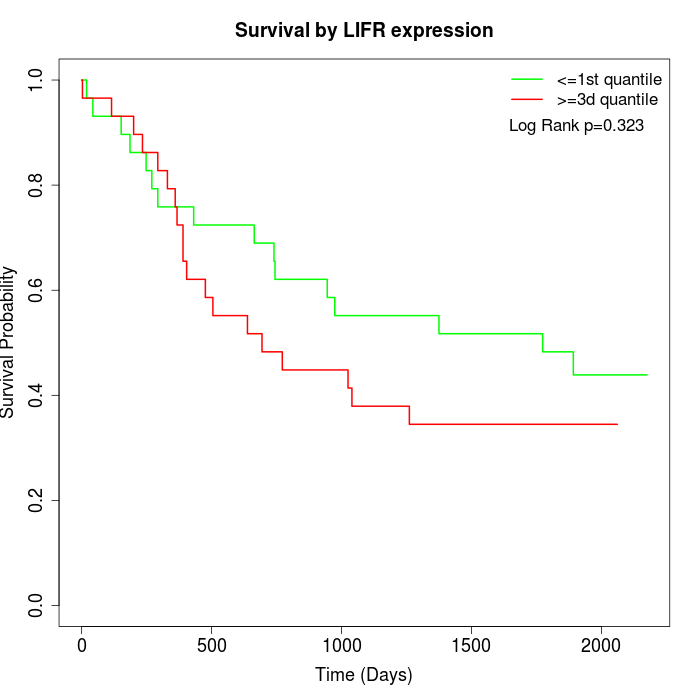
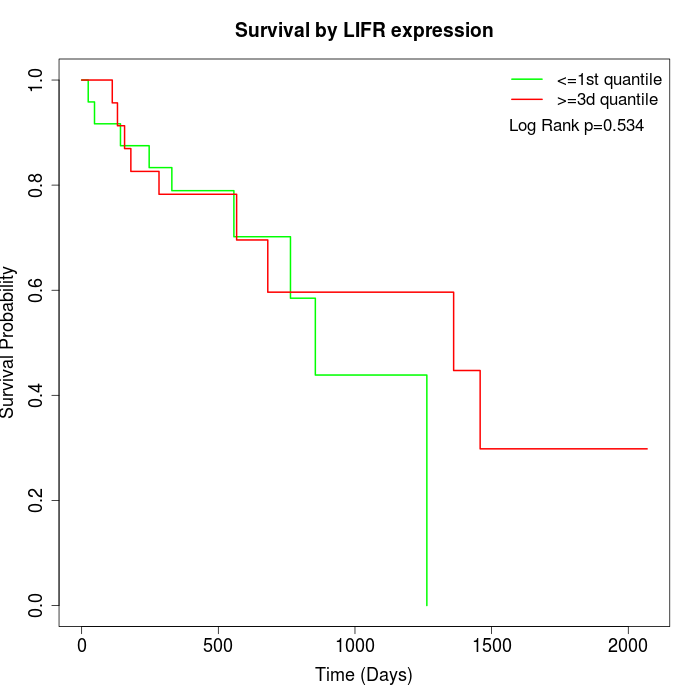
Survival by LIFR expression:
Note: Click image to view full size file.
Copy number change of LIFR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LIFR | 3977 | 10 | 0 | 20 | |
| GSE20123 | LIFR | 3977 | 10 | 0 | 20 | |
| GSE43470 | LIFR | 3977 | 18 | 0 | 25 | |
| GSE46452 | LIFR | 3977 | 5 | 22 | 32 | |
| GSE47630 | LIFR | 3977 | 6 | 12 | 22 | |
| GSE54993 | LIFR | 3977 | 5 | 4 | 61 | |
| GSE54994 | LIFR | 3977 | 25 | 2 | 26 | |
| GSE60625 | LIFR | 3977 | 0 | 0 | 11 | |
| GSE74703 | LIFR | 3977 | 14 | 0 | 22 | |
| GSE74704 | LIFR | 3977 | 9 | 0 | 11 | |
| TCGA | LIFR | 3977 | 53 | 5 | 38 |
Total number of gains: 155; Total number of losses: 45; Total Number of normals: 288.
Somatic mutations of LIFR:
Generating mutation plots.
Highly correlated genes for LIFR:
Showing top 20/1112 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LIFR | FAM13C | 0.817563 | 3 | 0 | 3 |
| LIFR | KCNE3 | 0.810465 | 3 | 0 | 3 |
| LIFR | ASPA | 0.797501 | 6 | 0 | 6 |
| LIFR | PREX2 | 0.790085 | 4 | 0 | 4 |
| LIFR | ABCA8 | 0.788101 | 6 | 0 | 6 |
| LIFR | C1QTNF7 | 0.782966 | 5 | 0 | 5 |
| LIFR | PDE1A | 0.780643 | 6 | 0 | 6 |
| LIFR | MYOCD | 0.777937 | 4 | 0 | 4 |
| LIFR | TCF21 | 0.776643 | 6 | 0 | 6 |
| LIFR | TMEM100 | 0.776366 | 5 | 0 | 5 |
| LIFR | MYRIP | 0.776124 | 6 | 0 | 6 |
| LIFR | RBPMS2 | 0.775888 | 5 | 0 | 5 |
| LIFR | SLIT3 | 0.773967 | 4 | 0 | 4 |
| LIFR | OGN | 0.771899 | 6 | 0 | 6 |
| LIFR | RCAN2 | 0.766597 | 6 | 0 | 6 |
| LIFR | RGN | 0.759702 | 6 | 0 | 6 |
| LIFR | DDAH1 | 0.759322 | 5 | 0 | 5 |
| LIFR | PEG3 | 0.757593 | 4 | 0 | 4 |
| LIFR | FAT4 | 0.755398 | 3 | 0 | 3 |
| LIFR | PDK2 | 0.755249 | 3 | 0 | 3 |
For details and further investigation, click here