| Full name: long intergenic non-protein coding RNA 689 | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 7q36.3 | ||
| Entrez ID: 154822 | HGNC ID: HGNC:27217 | Ensembl Gene: ENSG00000231419 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LINC00689:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LINC00689 | 154822 | 243261_at | -0.0858 | 0.7098 | |
| GSE26886 | LINC00689 | 154822 | 243261_at | -0.0959 | 0.5593 | |
| GSE45670 | LINC00689 | 154822 | 243261_at | -0.0268 | 0.8533 | |
| GSE53622 | LINC00689 | 154822 | 93201 | -0.1253 | 0.0632 | |
| GSE53624 | LINC00689 | 154822 | 93201 | -0.2003 | 0.0239 | |
| GSE63941 | LINC00689 | 154822 | 243261_at | 0.2316 | 0.2936 | |
| GSE77861 | LINC00689 | 154822 | 243261_at | -0.2014 | 0.0673 | |
| SRP133303 | LINC00689 | 154822 | RNAseq | -0.2993 | 0.2775 |
Upregulated datasets: 0; Downregulated datasets: 0.
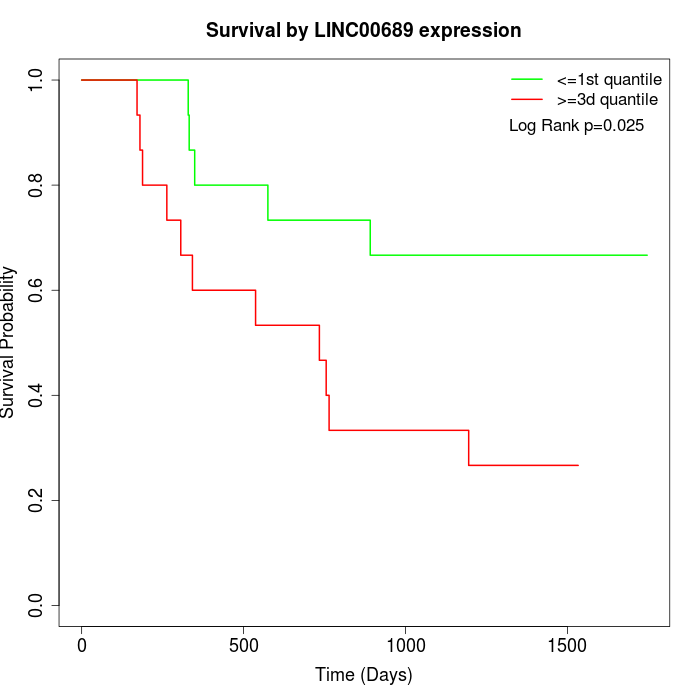
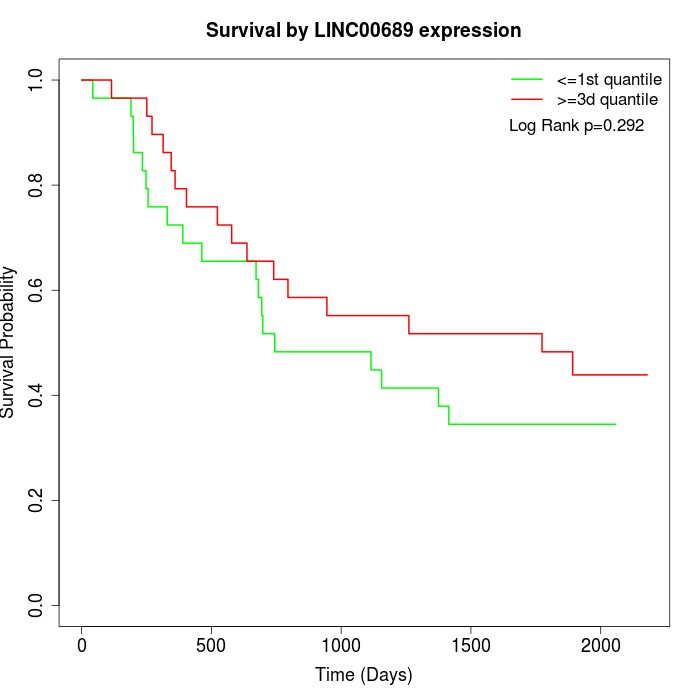
Survival by LINC00689 expression:
Note: Click image to view full size file.
Copy number change of LINC00689:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LINC00689 | 154822 | 3 | 3 | 24 | |
| GSE20123 | LINC00689 | 154822 | 2 | 3 | 25 | |
| GSE43470 | LINC00689 | 154822 | 2 | 5 | 36 | |
| GSE46452 | LINC00689 | 154822 | 7 | 2 | 50 | |
| GSE47630 | LINC00689 | 154822 | 6 | 9 | 25 | |
| GSE54993 | LINC00689 | 154822 | 3 | 3 | 64 | |
| GSE54994 | LINC00689 | 154822 | 8 | 9 | 36 | |
| GSE60625 | LINC00689 | 154822 | 0 | 0 | 11 | |
| GSE74703 | LINC00689 | 154822 | 2 | 4 | 30 | |
| GSE74704 | LINC00689 | 154822 | 1 | 3 | 16 | |
| TCGA | LINC00689 | 154822 | 28 | 26 | 42 |
Total number of gains: 62; Total number of losses: 67; Total Number of normals: 359.
Somatic mutations of LINC00689:
Generating mutation plots.
Highly correlated genes for LINC00689:
Showing top 20/164 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LINC00689 | CSNK1G2-AS1 | 0.724157 | 3 | 0 | 3 |
| LINC00689 | TSPAN11 | 0.703223 | 4 | 0 | 4 |
| LINC00689 | TCTA | 0.690896 | 3 | 0 | 3 |
| LINC00689 | FGA | 0.677607 | 3 | 0 | 3 |
| LINC00689 | CHRNA2 | 0.648521 | 5 | 0 | 4 |
| LINC00689 | LINC00315 | 0.647581 | 6 | 0 | 5 |
| LINC00689 | BTNL9 | 0.646452 | 3 | 0 | 3 |
| LINC00689 | PTH2 | 0.643488 | 4 | 0 | 3 |
| LINC00689 | SAMD4A | 0.64304 | 3 | 0 | 3 |
| LINC00689 | CD33 | 0.639241 | 3 | 0 | 3 |
| LINC00689 | KCNIP1 | 0.637441 | 3 | 0 | 3 |
| LINC00689 | FAM214B | 0.635357 | 3 | 0 | 3 |
| LINC00689 | UTF1 | 0.630927 | 4 | 0 | 3 |
| LINC00689 | CC2D2B | 0.627898 | 3 | 0 | 3 |
| LINC00689 | TAC3 | 0.626582 | 4 | 0 | 3 |
| LINC00689 | ENPP3 | 0.618918 | 3 | 0 | 3 |
| LINC00689 | CD1C | 0.615088 | 3 | 0 | 3 |
| LINC00689 | ABCA4 | 0.612313 | 3 | 0 | 3 |
| LINC00689 | CARD11 | 0.611507 | 3 | 0 | 3 |
| LINC00689 | SDK2 | 0.611406 | 4 | 0 | 3 |
For details and further investigation, click here