| Full name: T cell leukemia translocation altered | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 3p21.31 | ||
| Entrez ID: 6988 | HGNC ID: HGNC:11692 | Ensembl Gene: ENSG00000145022 | OMIM ID: 600690 |
| Drug and gene relationship at DGIdb | |||
Expression of TCTA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TCTA | 6988 | 203054_s_at | -0.3427 | 0.5049 | |
| GSE20347 | TCTA | 6988 | 203054_s_at | -0.2533 | 0.0261 | |
| GSE23400 | TCTA | 6988 | 203054_s_at | -0.2141 | 0.0000 | |
| GSE26886 | TCTA | 6988 | 203054_s_at | -0.5675 | 0.0014 | |
| GSE29001 | TCTA | 6988 | 203054_s_at | 0.0970 | 0.6800 | |
| GSE38129 | TCTA | 6988 | 203054_s_at | -0.3813 | 0.0016 | |
| GSE45670 | TCTA | 6988 | 203054_s_at | -0.0880 | 0.5155 | |
| GSE53622 | TCTA | 6988 | 101947 | -0.1414 | 0.0847 | |
| GSE53624 | TCTA | 6988 | 101947 | -0.2060 | 0.0038 | |
| GSE63941 | TCTA | 6988 | 203054_s_at | -0.9676 | 0.0498 | |
| GSE77861 | TCTA | 6988 | 203054_s_at | -0.2051 | 0.4084 | |
| GSE97050 | TCTA | 6988 | A_23_P58002 | -0.1386 | 0.5492 | |
| SRP007169 | TCTA | 6988 | RNAseq | -0.5657 | 0.1096 | |
| SRP008496 | TCTA | 6988 | RNAseq | -0.5098 | 0.0174 | |
| SRP064894 | TCTA | 6988 | RNAseq | 0.0623 | 0.7653 | |
| SRP133303 | TCTA | 6988 | RNAseq | -0.1751 | 0.1871 | |
| SRP159526 | TCTA | 6988 | RNAseq | -0.2843 | 0.1874 | |
| SRP193095 | TCTA | 6988 | RNAseq | -0.3802 | 0.0286 | |
| SRP219564 | TCTA | 6988 | RNAseq | -0.3363 | 0.4536 | |
| TCGA | TCTA | 6988 | RNAseq | -0.2760 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
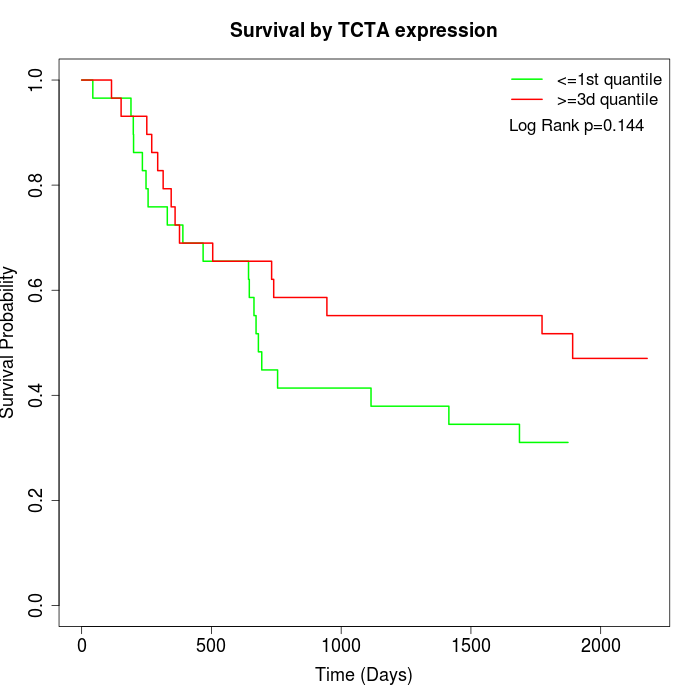
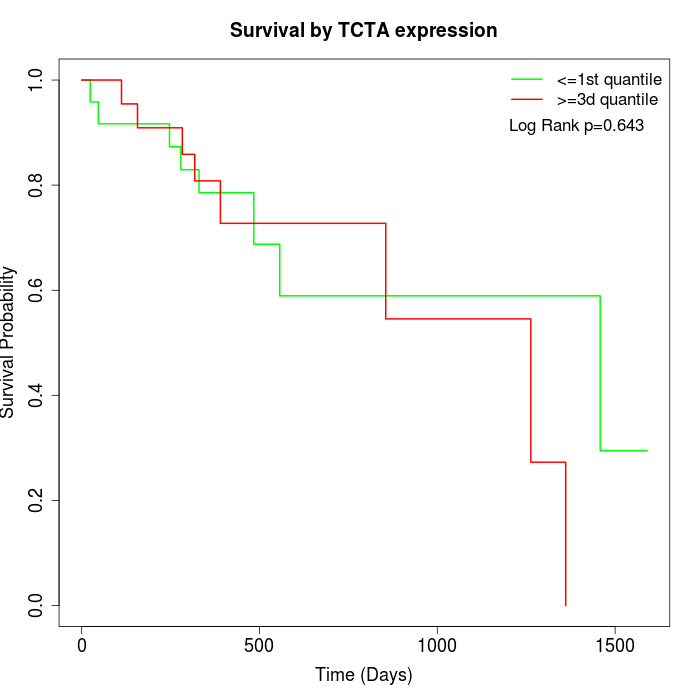
Survival by TCTA expression:
Note: Click image to view full size file.
Copy number change of TCTA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TCTA | 6988 | 0 | 18 | 12 | |
| GSE20123 | TCTA | 6988 | 0 | 19 | 11 | |
| GSE43470 | TCTA | 6988 | 0 | 18 | 25 | |
| GSE46452 | TCTA | 6988 | 2 | 16 | 41 | |
| GSE47630 | TCTA | 6988 | 1 | 24 | 15 | |
| GSE54993 | TCTA | 6988 | 6 | 2 | 62 | |
| GSE54994 | TCTA | 6988 | 0 | 36 | 17 | |
| GSE60625 | TCTA | 6988 | 5 | 0 | 6 | |
| GSE74703 | TCTA | 6988 | 0 | 14 | 22 | |
| GSE74704 | TCTA | 6988 | 0 | 12 | 8 | |
| TCGA | TCTA | 6988 | 0 | 78 | 18 |
Total number of gains: 14; Total number of losses: 237; Total Number of normals: 237.
Somatic mutations of TCTA:
Generating mutation plots.
Highly correlated genes for TCTA:
Showing top 20/250 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TCTA | RNF180 | 0.743177 | 3 | 0 | 3 |
| TCTA | KIRREL2 | 0.743161 | 3 | 0 | 3 |
| TCTA | IL11RA | 0.717924 | 3 | 0 | 3 |
| TCTA | VPS26B | 0.717702 | 4 | 0 | 4 |
| TCTA | SMPX | 0.699318 | 4 | 0 | 3 |
| TCTA | LINC00689 | 0.690896 | 3 | 0 | 3 |
| TCTA | SCARA5 | 0.689898 | 3 | 0 | 3 |
| TCTA | ANKRD44 | 0.689418 | 3 | 0 | 3 |
| TCTA | MYOM1 | 0.68914 | 4 | 0 | 4 |
| TCTA | SYCE1 | 0.684478 | 3 | 0 | 3 |
| TCTA | L1CAM | 0.670939 | 4 | 0 | 3 |
| TCTA | RNF38 | 0.664239 | 4 | 0 | 3 |
| TCTA | LINC01300 | 0.660663 | 3 | 0 | 3 |
| TCTA | SCRG1 | 0.647237 | 4 | 0 | 4 |
| TCTA | GNAZ | 0.64704 | 5 | 0 | 5 |
| TCTA | SGCD | 0.645271 | 4 | 0 | 3 |
| TCTA | ANK2 | 0.645208 | 4 | 0 | 3 |
| TCTA | CLIP3 | 0.641494 | 5 | 0 | 4 |
| TCTA | BACE1 | 0.640416 | 4 | 0 | 3 |
| TCTA | FGL2 | 0.639628 | 4 | 0 | 3 |
For details and further investigation, click here