| Full name: lipase C, hepatic type | Alias Symbol: HL|HTGL | ||
| Type: protein-coding gene | Cytoband: 15q21.3 | ||
| Entrez ID: 3990 | HGNC ID: HGNC:6619 | Ensembl Gene: ENSG00000166035 | OMIM ID: 151670 |
| Drug and gene relationship at DGIdb | |||
Expression of LIPC:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LIPC | 3990 | 206606_at | -0.2814 | 0.3485 | |
| GSE20347 | LIPC | 3990 | 206606_at | -0.1012 | 0.1083 | |
| GSE23400 | LIPC | 3990 | 206606_at | -0.1506 | 0.0000 | |
| GSE26886 | LIPC | 3990 | 206606_at | -0.1742 | 0.1007 | |
| GSE29001 | LIPC | 3990 | 206606_at | -0.0858 | 0.6079 | |
| GSE38129 | LIPC | 3990 | 206606_at | -0.2130 | 0.0041 | |
| GSE45670 | LIPC | 3990 | 206606_at | -0.1908 | 0.1500 | |
| GSE53622 | LIPC | 3990 | 67832 | -0.8368 | 0.0000 | |
| GSE53624 | LIPC | 3990 | 67832 | -0.5688 | 0.0036 | |
| GSE63941 | LIPC | 3990 | 206606_at | -0.1774 | 0.2465 | |
| GSE77861 | LIPC | 3990 | 206606_at | -0.1430 | 0.0668 | |
| SRP133303 | LIPC | 3990 | RNAseq | 0.1070 | 0.6787 | |
| TCGA | LIPC | 3990 | RNAseq | -1.4223 | 0.0143 |
Upregulated datasets: 0; Downregulated datasets: 1.
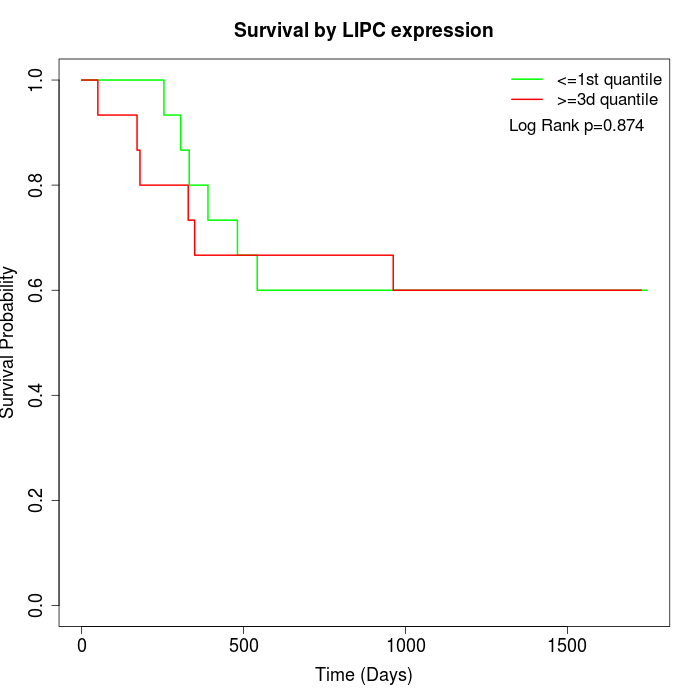
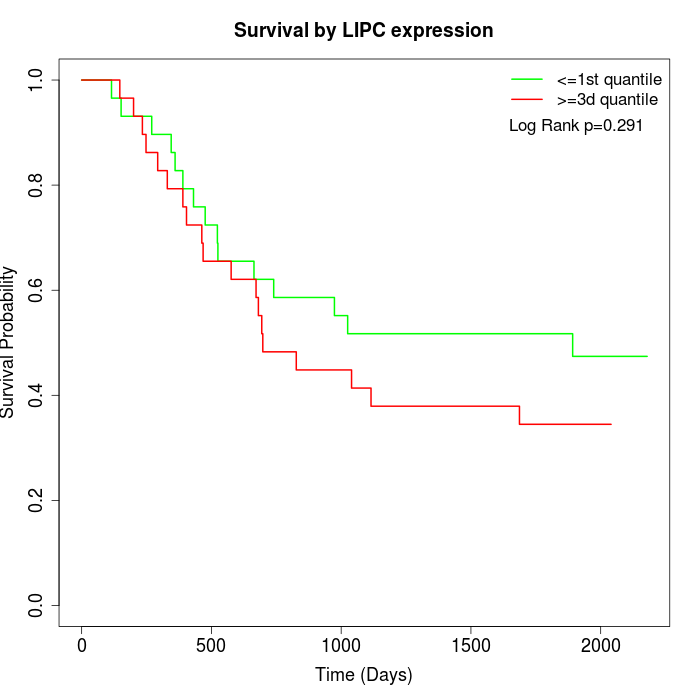
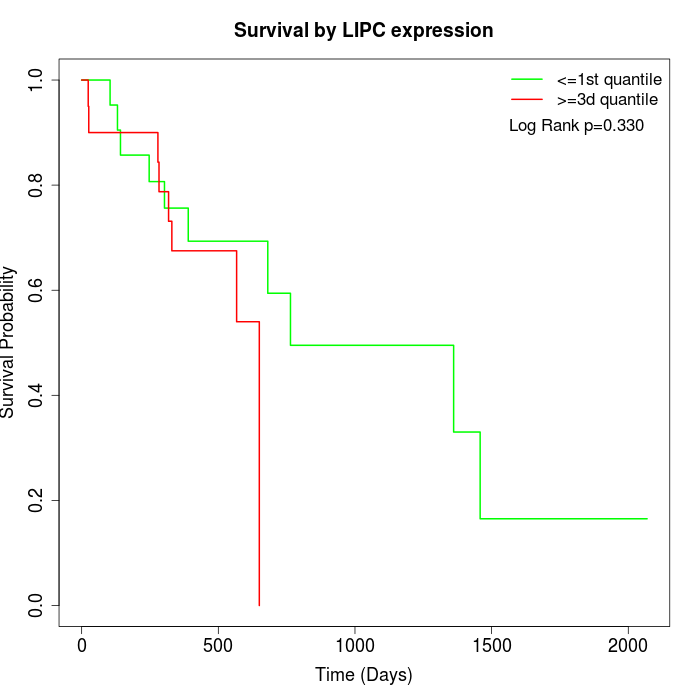
Survival by LIPC expression:
Note: Click image to view full size file.
Copy number change of LIPC:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LIPC | 3990 | 6 | 4 | 20 | |
| GSE20123 | LIPC | 3990 | 6 | 3 | 21 | |
| GSE43470 | LIPC | 3990 | 4 | 4 | 35 | |
| GSE46452 | LIPC | 3990 | 3 | 7 | 49 | |
| GSE47630 | LIPC | 3990 | 8 | 10 | 22 | |
| GSE54993 | LIPC | 3990 | 5 | 6 | 59 | |
| GSE54994 | LIPC | 3990 | 6 | 8 | 39 | |
| GSE60625 | LIPC | 3990 | 4 | 0 | 7 | |
| GSE74703 | LIPC | 3990 | 4 | 3 | 29 | |
| GSE74704 | LIPC | 3990 | 2 | 3 | 15 | |
| TCGA | LIPC | 3990 | 10 | 17 | 69 |
Total number of gains: 58; Total number of losses: 65; Total Number of normals: 365.
Somatic mutations of LIPC:
Generating mutation plots.
Highly correlated genes for LIPC:
Showing top 20/465 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LIPC | NR6A1 | 0.712688 | 4 | 0 | 4 |
| LIPC | MTAP | 0.702121 | 3 | 0 | 3 |
| LIPC | GUCY2C | 0.690872 | 3 | 0 | 3 |
| LIPC | PF4 | 0.686027 | 6 | 0 | 5 |
| LIPC | P2RX6 | 0.674309 | 4 | 0 | 4 |
| LIPC | GREB1L | 0.672901 | 3 | 0 | 3 |
| LIPC | ODF4 | 0.67174 | 3 | 0 | 3 |
| LIPC | BMP15 | 0.671018 | 3 | 0 | 3 |
| LIPC | PSG7 | 0.6618 | 4 | 0 | 4 |
| LIPC | BCL2L15 | 0.659858 | 3 | 0 | 3 |
| LIPC | TRIM3 | 0.659195 | 4 | 0 | 4 |
| LIPC | TGM4 | 0.65681 | 5 | 0 | 4 |
| LIPC | ADAM18 | 0.65444 | 3 | 0 | 3 |
| LIPC | SEMA3B | 0.654058 | 4 | 0 | 4 |
| LIPC | CSN2 | 0.653311 | 3 | 0 | 3 |
| LIPC | ZG16 | 0.648751 | 3 | 0 | 3 |
| LIPC | SNTG1 | 0.648179 | 3 | 0 | 3 |
| LIPC | GABRA1 | 0.646447 | 3 | 0 | 3 |
| LIPC | ATP1A1-AS1 | 0.646442 | 3 | 0 | 3 |
| LIPC | C22orf31 | 0.64595 | 4 | 0 | 3 |
For details and further investigation, click here