| Full name: leucine rich repeat neuronal 1 | Alias Symbol: FIGLER3 | ||
| Type: protein-coding gene | Cytoband: 3p26.2 | ||
| Entrez ID: 57633 | HGNC ID: HGNC:20980 | Ensembl Gene: ENSG00000175928 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LRRN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LRRN1 | 57633 | 226884_at | -0.2797 | 0.4368 | |
| GSE26886 | LRRN1 | 57633 | 226884_at | 0.1698 | 0.5095 | |
| GSE45670 | LRRN1 | 57633 | 226884_at | -0.3073 | 0.1977 | |
| GSE53622 | LRRN1 | 57633 | 61552 | -0.1347 | 0.6631 | |
| GSE53624 | LRRN1 | 57633 | 61552 | -0.2703 | 0.1965 | |
| GSE63941 | LRRN1 | 57633 | 226884_at | -1.4090 | 0.1613 | |
| GSE77861 | LRRN1 | 57633 | 226884_at | -0.0607 | 0.4728 | |
| GSE97050 | LRRN1 | 57633 | A_24_P240187 | 0.3840 | 0.3240 | |
| SRP133303 | LRRN1 | 57633 | RNAseq | 0.1092 | 0.8168 | |
| SRP159526 | LRRN1 | 57633 | RNAseq | 2.8755 | 0.0022 | |
| SRP219564 | LRRN1 | 57633 | RNAseq | 0.1859 | 0.8229 | |
| TCGA | LRRN1 | 57633 | RNAseq | -0.7231 | 0.0759 |
Upregulated datasets: 1; Downregulated datasets: 0.
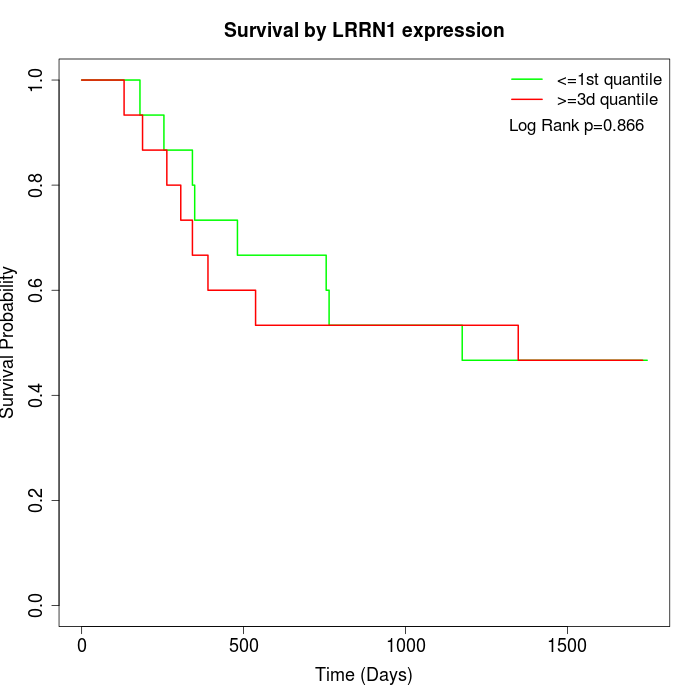
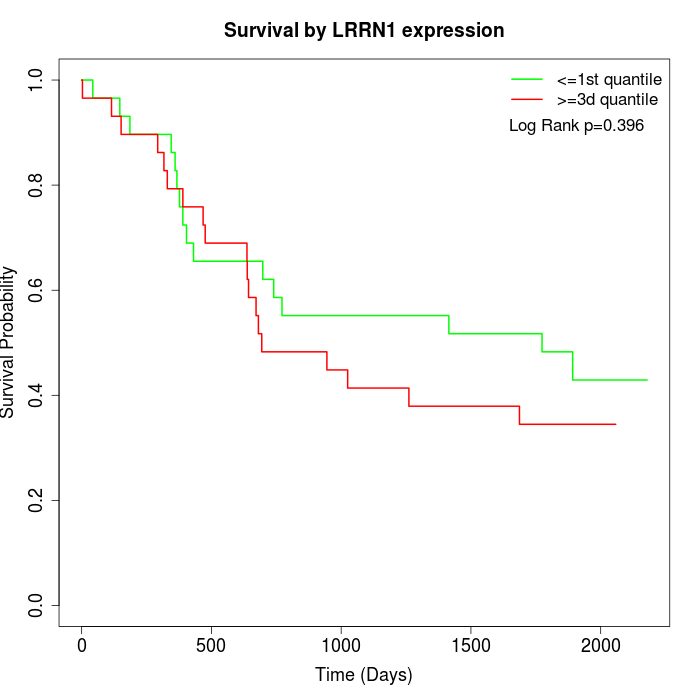
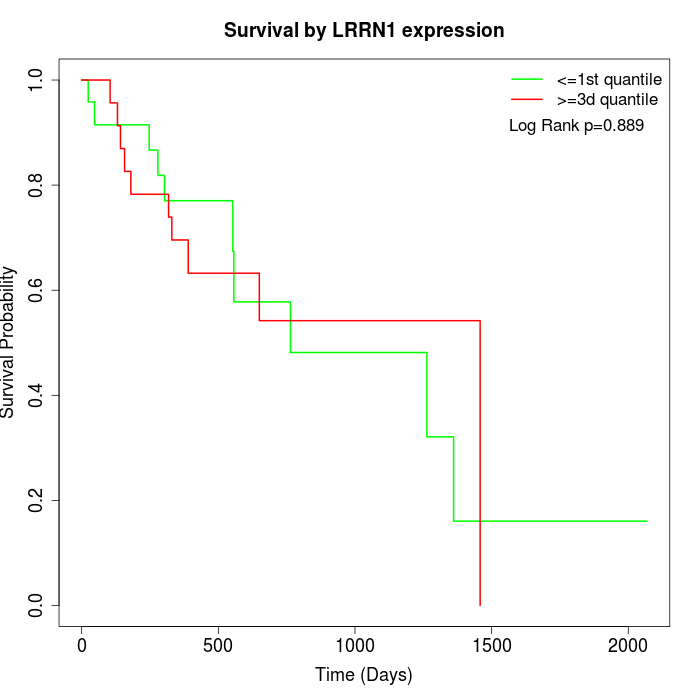
Survival by LRRN1 expression:
Note: Click image to view full size file.
Copy number change of LRRN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LRRN1 | 57633 | 1 | 18 | 11 | |
| GSE20123 | LRRN1 | 57633 | 1 | 18 | 11 | |
| GSE43470 | LRRN1 | 57633 | 0 | 21 | 22 | |
| GSE46452 | LRRN1 | 57633 | 2 | 18 | 39 | |
| GSE47630 | LRRN1 | 57633 | 1 | 24 | 15 | |
| GSE54993 | LRRN1 | 57633 | 6 | 1 | 63 | |
| GSE54994 | LRRN1 | 57633 | 1 | 32 | 20 | |
| GSE60625 | LRRN1 | 57633 | 5 | 0 | 6 | |
| GSE74703 | LRRN1 | 57633 | 0 | 18 | 18 | |
| GSE74704 | LRRN1 | 57633 | 1 | 11 | 8 | |
| TCGA | LRRN1 | 57633 | 2 | 67 | 27 |
Total number of gains: 20; Total number of losses: 228; Total Number of normals: 240.
Somatic mutations of LRRN1:
Generating mutation plots.
Highly correlated genes for LRRN1:
Showing top 20/71 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LRRN1 | ARSG | 0.745927 | 3 | 0 | 3 |
| LRRN1 | ZNF248 | 0.672681 | 3 | 0 | 3 |
| LRRN1 | MPI | 0.661273 | 3 | 0 | 3 |
| LRRN1 | KRT33B | 0.658056 | 3 | 0 | 3 |
| LRRN1 | TPCN1 | 0.652412 | 3 | 0 | 3 |
| LRRN1 | RNF165 | 0.651681 | 3 | 0 | 3 |
| LRRN1 | PKP2 | 0.64018 | 3 | 0 | 3 |
| LRRN1 | PDE8B | 0.628197 | 4 | 0 | 3 |
| LRRN1 | MLLT3 | 0.619301 | 4 | 0 | 4 |
| LRRN1 | ZNF414 | 0.61528 | 3 | 0 | 3 |
| LRRN1 | CSGALNACT1 | 0.607788 | 4 | 0 | 3 |
| LRRN1 | GCKR | 0.606824 | 3 | 0 | 3 |
| LRRN1 | ARSD | 0.600905 | 3 | 0 | 3 |
| LRRN1 | STK33 | 0.600865 | 4 | 0 | 3 |
| LRRN1 | GIPC2 | 0.595586 | 3 | 0 | 3 |
| LRRN1 | CLK3 | 0.594661 | 3 | 0 | 3 |
| LRRN1 | SLC18A2 | 0.588082 | 4 | 0 | 3 |
| LRRN1 | DDAH1 | 0.587718 | 3 | 0 | 3 |
| LRRN1 | CDK19 | 0.586736 | 3 | 0 | 3 |
| LRRN1 | GBGT1 | 0.578065 | 4 | 0 | 3 |
For details and further investigation, click here