| Full name: lysozyme like 1 | Alias Symbol: MGC33408|LYC2 | ||
| Type: protein-coding gene | Cytoband: 10p12.1-p11.23 | ||
| Entrez ID: 84569 | HGNC ID: HGNC:30502 | Ensembl Gene: ENSG00000120563 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LYZL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LYZL1 | 84569 | 233973_at | 0.0595 | 0.7934 | |
| GSE26886 | LYZL1 | 84569 | 233973_at | 0.1564 | 0.0189 | |
| GSE45670 | LYZL1 | 84569 | 233973_at | 0.1164 | 0.1166 | |
| GSE53622 | LYZL1 | 84569 | 86138 | 0.4408 | 0.0690 | |
| GSE53624 | LYZL1 | 84569 | 86138 | 0.2097 | 0.1872 | |
| GSE63941 | LYZL1 | 84569 | 233973_at | -0.1187 | 0.4419 | |
| GSE77861 | LYZL1 | 84569 | 233973_at | -0.0582 | 0.5949 |
Upregulated datasets: 0; Downregulated datasets: 0.
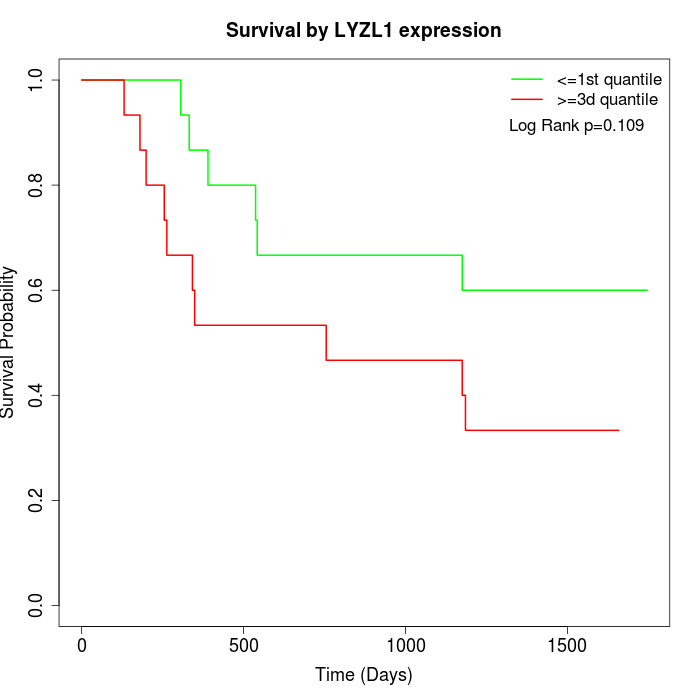
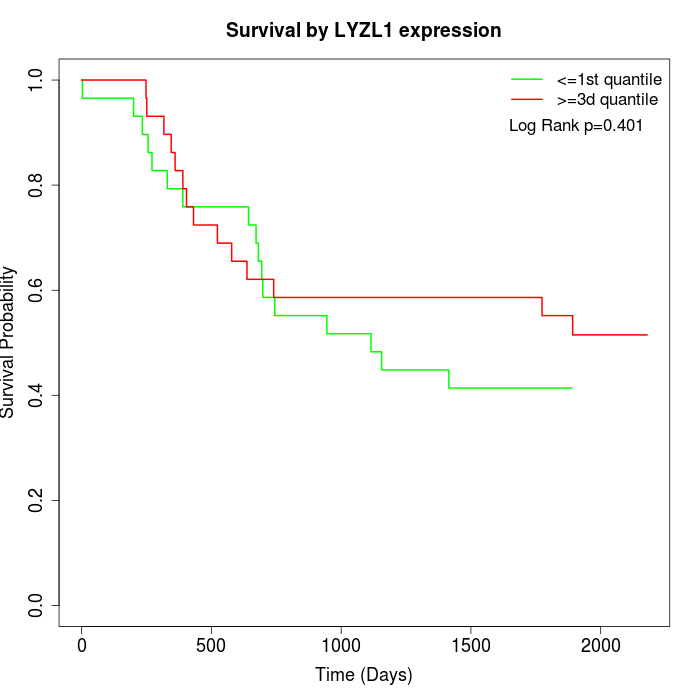
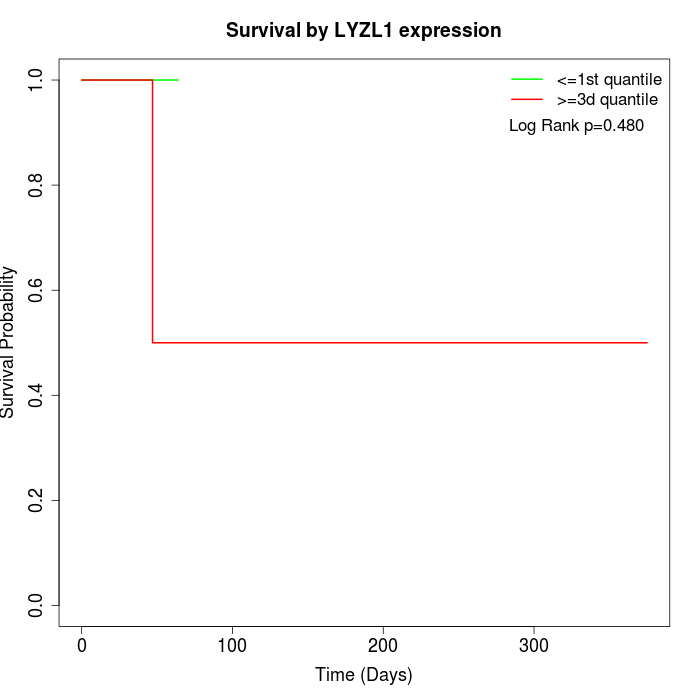
Survival by LYZL1 expression:
Note: Click image to view full size file.
Copy number change of LYZL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LYZL1 | 84569 | 5 | 7 | 18 | |
| GSE20123 | LYZL1 | 84569 | 5 | 6 | 19 | |
| GSE43470 | LYZL1 | 84569 | 3 | 4 | 36 | |
| GSE46452 | LYZL1 | 84569 | 1 | 14 | 44 | |
| GSE47630 | LYZL1 | 84569 | 5 | 15 | 20 | |
| GSE54993 | LYZL1 | 84569 | 9 | 0 | 61 | |
| GSE54994 | LYZL1 | 84569 | 3 | 9 | 41 | |
| GSE60625 | LYZL1 | 84569 | 0 | 0 | 11 | |
| GSE74703 | LYZL1 | 84569 | 2 | 2 | 32 | |
| GSE74704 | LYZL1 | 84569 | 0 | 5 | 15 | |
| TCGA | LYZL1 | 84569 | 19 | 23 | 54 |
Total number of gains: 52; Total number of losses: 85; Total Number of normals: 351.
Somatic mutations of LYZL1:
Generating mutation plots.
Highly correlated genes for LYZL1:
Showing top 20/58 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LYZL1 | LINC01281 | 0.676271 | 3 | 0 | 3 |
| LYZL1 | RPGRIP1L | 0.674485 | 3 | 0 | 3 |
| LYZL1 | USP2-AS1 | 0.672831 | 3 | 0 | 3 |
| LYZL1 | ABCA13 | 0.670296 | 3 | 0 | 3 |
| LYZL1 | TMEM105 | 0.669808 | 3 | 0 | 3 |
| LYZL1 | LINC01350 | 0.654312 | 3 | 0 | 3 |
| LYZL1 | SLC52A1 | 0.653012 | 3 | 0 | 3 |
| LYZL1 | FNDC9 | 0.65249 | 3 | 0 | 3 |
| LYZL1 | CCDC33 | 0.649633 | 3 | 0 | 3 |
| LYZL1 | BTNL2 | 0.647514 | 3 | 0 | 3 |
| LYZL1 | LINC00857 | 0.640209 | 3 | 0 | 3 |
| LYZL1 | FOXE3 | 0.639454 | 4 | 0 | 4 |
| LYZL1 | ZSCAN16-AS1 | 0.634443 | 3 | 0 | 3 |
| LYZL1 | KRTAP4-8 | 0.627559 | 3 | 0 | 3 |
| LYZL1 | CASS4 | 0.623355 | 3 | 0 | 3 |
| LYZL1 | GRIK4 | 0.623083 | 3 | 0 | 3 |
| LYZL1 | LINC00051 | 0.620476 | 3 | 0 | 3 |
| LYZL1 | CYP11B1 | 0.614561 | 3 | 0 | 3 |
| LYZL1 | HGFAC | 0.612126 | 3 | 0 | 3 |
| LYZL1 | FAM205A | 0.611293 | 3 | 0 | 3 |
For details and further investigation, click here