| Full name: mitogen-activated protein kinase kinase kinase 3 | Alias Symbol: MAPKKK3 | ||
| Type: protein-coding gene | Cytoband: 17q23.3 | ||
| Entrez ID: 4215 | HGNC ID: HGNC:6855 | Ensembl Gene: ENSG00000198909 | OMIM ID: 602539 |
| Drug and gene relationship at DGIdb | |||
MAP3K3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04722 | Neurotrophin signaling pathway | |
| hsa04912 | GnRH signaling pathway | |
| hsa05166 | HTLV-I infection |
Expression of MAP3K3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP3K3 | 4215 | 203514_at | -0.3007 | 0.3206 | |
| GSE20347 | MAP3K3 | 4215 | 203514_at | -0.2628 | 0.0136 | |
| GSE23400 | MAP3K3 | 4215 | 203514_at | -0.2952 | 0.0000 | |
| GSE26886 | MAP3K3 | 4215 | 203514_at | -0.6069 | 0.0005 | |
| GSE29001 | MAP3K3 | 4215 | 203514_at | -0.5181 | 0.0046 | |
| GSE38129 | MAP3K3 | 4215 | 203514_at | -0.4085 | 0.0221 | |
| GSE45670 | MAP3K3 | 4215 | 203514_at | -0.1886 | 0.0244 | |
| GSE53622 | MAP3K3 | 4215 | 55879 | -0.6546 | 0.0000 | |
| GSE53624 | MAP3K3 | 4215 | 55879 | -0.3957 | 0.0000 | |
| GSE63941 | MAP3K3 | 4215 | 227131_at | -1.2137 | 0.0240 | |
| GSE77861 | MAP3K3 | 4215 | 203514_at | -0.1165 | 0.4060 | |
| GSE97050 | MAP3K3 | 4215 | A_33_P3271430 | -0.3333 | 0.2151 | |
| SRP007169 | MAP3K3 | 4215 | RNAseq | 0.3584 | 0.4518 | |
| SRP008496 | MAP3K3 | 4215 | RNAseq | 0.8633 | 0.0322 | |
| SRP064894 | MAP3K3 | 4215 | RNAseq | 0.0447 | 0.8211 | |
| SRP133303 | MAP3K3 | 4215 | RNAseq | -0.3282 | 0.0807 | |
| SRP159526 | MAP3K3 | 4215 | RNAseq | -0.3270 | 0.0761 | |
| SRP193095 | MAP3K3 | 4215 | RNAseq | -0.0445 | 0.6666 | |
| SRP219564 | MAP3K3 | 4215 | RNAseq | -0.1339 | 0.6710 | |
| TCGA | MAP3K3 | 4215 | RNAseq | -0.2065 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 1.
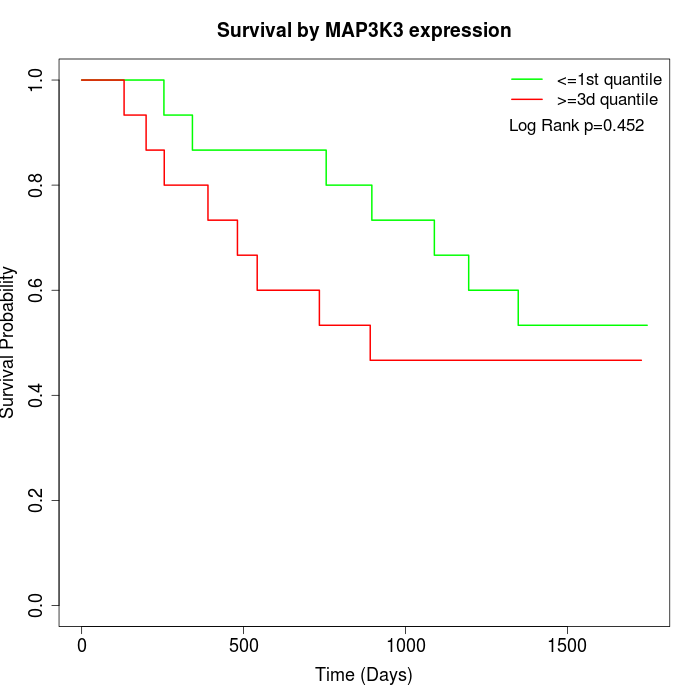
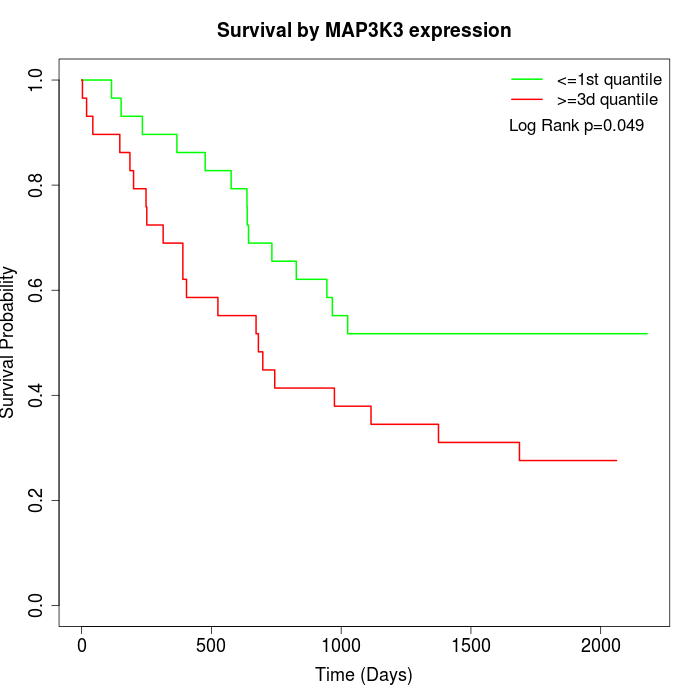
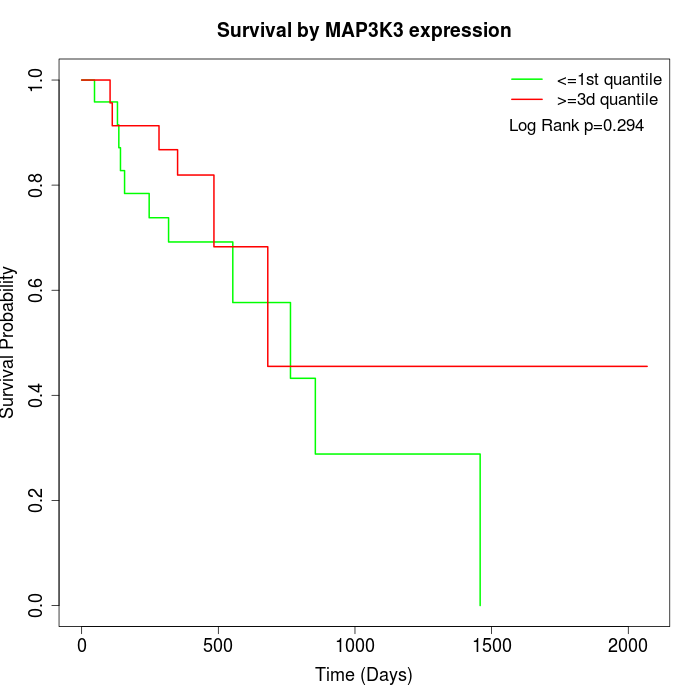
Survival by MAP3K3 expression:
Note: Click image to view full size file.
Copy number change of MAP3K3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP3K3 | 4215 | 3 | 1 | 26 | |
| GSE20123 | MAP3K3 | 4215 | 3 | 1 | 26 | |
| GSE43470 | MAP3K3 | 4215 | 4 | 0 | 39 | |
| GSE46452 | MAP3K3 | 4215 | 31 | 0 | 28 | |
| GSE47630 | MAP3K3 | 4215 | 7 | 1 | 32 | |
| GSE54993 | MAP3K3 | 4215 | 2 | 5 | 63 | |
| GSE54994 | MAP3K3 | 4215 | 9 | 4 | 40 | |
| GSE60625 | MAP3K3 | 4215 | 4 | 0 | 7 | |
| GSE74703 | MAP3K3 | 4215 | 4 | 0 | 32 | |
| GSE74704 | MAP3K3 | 4215 | 3 | 1 | 16 | |
| TCGA | MAP3K3 | 4215 | 31 | 9 | 56 |
Total number of gains: 101; Total number of losses: 22; Total Number of normals: 365.
Somatic mutations of MAP3K3:
Generating mutation plots.
Highly correlated genes for MAP3K3:
Showing top 20/888 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP3K3 | USP9Y | 0.730333 | 3 | 0 | 3 |
| MAP3K3 | MSRB3 | 0.729653 | 4 | 0 | 4 |
| MAP3K3 | CLEC3B | 0.728203 | 3 | 0 | 3 |
| MAP3K3 | FIBIN | 0.725399 | 3 | 0 | 3 |
| MAP3K3 | ZNF582 | 0.716701 | 4 | 0 | 4 |
| MAP3K3 | TMOD1 | 0.71565 | 4 | 0 | 4 |
| MAP3K3 | RCSD1 | 0.708645 | 3 | 0 | 3 |
| MAP3K3 | GNPDA2 | 0.695675 | 3 | 0 | 3 |
| MAP3K3 | KCNK2 | 0.693615 | 3 | 0 | 3 |
| MAP3K3 | DNAJC15 | 0.688194 | 3 | 0 | 3 |
| MAP3K3 | PRELP | 0.687731 | 6 | 0 | 5 |
| MAP3K3 | TNFSF12 | 0.686424 | 3 | 0 | 3 |
| MAP3K3 | CYP21A2 | 0.683311 | 3 | 0 | 3 |
| MAP3K3 | LYVE1 | 0.681156 | 5 | 0 | 5 |
| MAP3K3 | TBC1D2B | 0.679452 | 3 | 0 | 3 |
| MAP3K3 | ZNF383 | 0.67621 | 3 | 0 | 3 |
| MAP3K3 | EVI5L | 0.67479 | 3 | 0 | 3 |
| MAP3K3 | LIMD2 | 0.674123 | 5 | 0 | 4 |
| MAP3K3 | GNG7 | 0.673239 | 7 | 0 | 7 |
| MAP3K3 | GAS7 | 0.671723 | 11 | 0 | 10 |
For details and further investigation, click here