| Full name: zinc finger protein 582 | Alias Symbol: FLJ30927 | ||
| Type: protein-coding gene | Cytoband: 19q13.43 | ||
| Entrez ID: 147948 | HGNC ID: HGNC:26421 | Ensembl Gene: ENSG00000018869 | OMIM ID: 615600 |
| Drug and gene relationship at DGIdb | |||
Expression of ZNF582:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ZNF582 | 147948 | 241602_at | -0.2746 | 0.2942 | |
| GSE26886 | ZNF582 | 147948 | 241602_at | -0.6683 | 0.0000 | |
| GSE45670 | ZNF582 | 147948 | 241602_at | -0.0333 | 0.8076 | |
| GSE53622 | ZNF582 | 147948 | 64655 | -1.6219 | 0.0000 | |
| GSE53624 | ZNF582 | 147948 | 64655 | -1.5691 | 0.0000 | |
| GSE63941 | ZNF582 | 147948 | 241602_at | -0.9801 | 0.0000 | |
| GSE77861 | ZNF582 | 147948 | 241602_at | -0.1989 | 0.1245 | |
| SRP133303 | ZNF582 | 147948 | RNAseq | -1.0618 | 0.0034 | |
| SRP159526 | ZNF582 | 147948 | RNAseq | -1.4851 | 0.0156 | |
| SRP193095 | ZNF582 | 147948 | RNAseq | -0.5929 | 0.0179 | |
| TCGA | ZNF582 | 147948 | RNAseq | -1.4816 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 5.
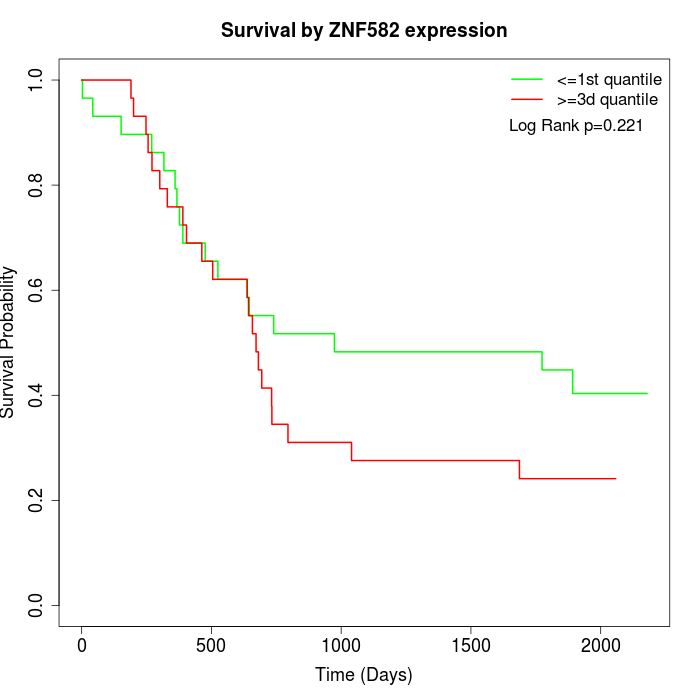
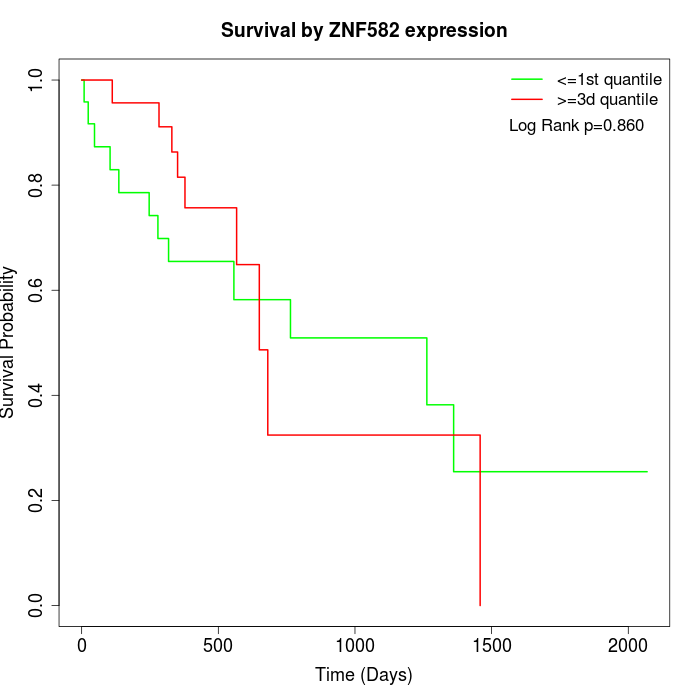
Survival by ZNF582 expression:
Note: Click image to view full size file.
Copy number change of ZNF582:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZNF582 | 147948 | 3 | 4 | 23 | |
| GSE20123 | ZNF582 | 147948 | 3 | 3 | 24 | |
| GSE43470 | ZNF582 | 147948 | 3 | 9 | 31 | |
| GSE46452 | ZNF582 | 147948 | 45 | 1 | 13 | |
| GSE47630 | ZNF582 | 147948 | 8 | 6 | 26 | |
| GSE54993 | ZNF582 | 147948 | 18 | 4 | 48 | |
| GSE54994 | ZNF582 | 147948 | 5 | 12 | 36 | |
| GSE60625 | ZNF582 | 147948 | 9 | 0 | 2 | |
| GSE74703 | ZNF582 | 147948 | 3 | 6 | 27 | |
| GSE74704 | ZNF582 | 147948 | 3 | 1 | 16 | |
| TCGA | ZNF582 | 147948 | 19 | 15 | 62 |
Total number of gains: 119; Total number of losses: 61; Total Number of normals: 308.
Somatic mutations of ZNF582:
Generating mutation plots.
Highly correlated genes for ZNF582:
Showing top 20/715 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZNF582 | TCF21 | 0.802974 | 4 | 0 | 4 |
| ZNF582 | BHMT2 | 0.791333 | 4 | 0 | 4 |
| ZNF582 | F10 | 0.780006 | 4 | 0 | 4 |
| ZNF582 | HSPB2 | 0.77512 | 4 | 0 | 4 |
| ZNF582 | NME5 | 0.757517 | 5 | 0 | 5 |
| ZNF582 | SHISA3 | 0.7563 | 4 | 0 | 4 |
| ZNF582 | RSPO2 | 0.756243 | 3 | 0 | 3 |
| ZNF582 | CADM3 | 0.754621 | 3 | 0 | 3 |
| ZNF582 | ZNF667-AS1 | 0.75336 | 5 | 0 | 5 |
| ZNF582 | FHL5 | 0.75127 | 3 | 0 | 3 |
| ZNF582 | ZEB2 | 0.748355 | 4 | 0 | 4 |
| ZNF582 | ZNF385D | 0.746515 | 3 | 0 | 3 |
| ZNF582 | AGTR1 | 0.746168 | 4 | 0 | 4 |
| ZNF582 | SYNE1 | 0.745143 | 4 | 0 | 4 |
| ZNF582 | PABPC5 | 0.74338 | 4 | 0 | 4 |
| ZNF582 | GRK5 | 0.741497 | 4 | 0 | 4 |
| ZNF582 | RUNX1T1 | 0.738865 | 3 | 0 | 3 |
| ZNF582 | FBXL7 | 0.735141 | 4 | 0 | 4 |
| ZNF582 | DPT | 0.734991 | 4 | 0 | 4 |
| ZNF582 | ASB5 | 0.732901 | 4 | 0 | 4 |
For details and further investigation, click here