| Full name: microtubule affinity regulating kinase 4 | Alias Symbol: Nbla00650|FLJ90097|KIAA1860|PAR-1D | ||
| Type: protein-coding gene | Cytoband: 19q13.32 | ||
| Entrez ID: 57787 | HGNC ID: HGNC:13538 | Ensembl Gene: ENSG00000007047 | OMIM ID: 606495 |
| Drug and gene relationship at DGIdb | |||
Expression of MARK4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MARK4 | 57787 | 55065_at | 0.5164 | 0.2628 | |
| GSE20347 | MARK4 | 57787 | 55065_at | 0.0775 | 0.5392 | |
| GSE23400 | MARK4 | 57787 | 221560_at | 0.0810 | 0.0960 | |
| GSE26886 | MARK4 | 57787 | 55065_at | 0.9283 | 0.0020 | |
| GSE29001 | MARK4 | 57787 | 55065_at | 0.0724 | 0.9048 | |
| GSE38129 | MARK4 | 57787 | 55065_at | 0.0558 | 0.6574 | |
| GSE45670 | MARK4 | 57787 | 55065_at | 0.0110 | 0.9481 | |
| GSE53622 | MARK4 | 57787 | 21752 | -0.1550 | 0.0050 | |
| GSE53624 | MARK4 | 57787 | 21752 | -0.3691 | 0.0000 | |
| GSE63941 | MARK4 | 57787 | 55065_at | -0.2267 | 0.5545 | |
| GSE77861 | MARK4 | 57787 | 221560_at | 0.2710 | 0.1375 | |
| GSE97050 | MARK4 | 57787 | A_33_P3322428 | 0.0725 | 0.7360 | |
| SRP007169 | MARK4 | 57787 | RNAseq | -0.1711 | 0.7023 | |
| SRP008496 | MARK4 | 57787 | RNAseq | -0.2000 | 0.5434 | |
| SRP064894 | MARK4 | 57787 | RNAseq | 0.2277 | 0.2431 | |
| SRP133303 | MARK4 | 57787 | RNAseq | 0.3719 | 0.0495 | |
| SRP159526 | MARK4 | 57787 | RNAseq | 0.7983 | 0.0057 | |
| SRP193095 | MARK4 | 57787 | RNAseq | 0.2372 | 0.0775 | |
| SRP219564 | MARK4 | 57787 | RNAseq | -0.2625 | 0.2397 | |
| TCGA | MARK4 | 57787 | RNAseq | 0.0557 | 0.2503 |
Upregulated datasets: 0; Downregulated datasets: 0.
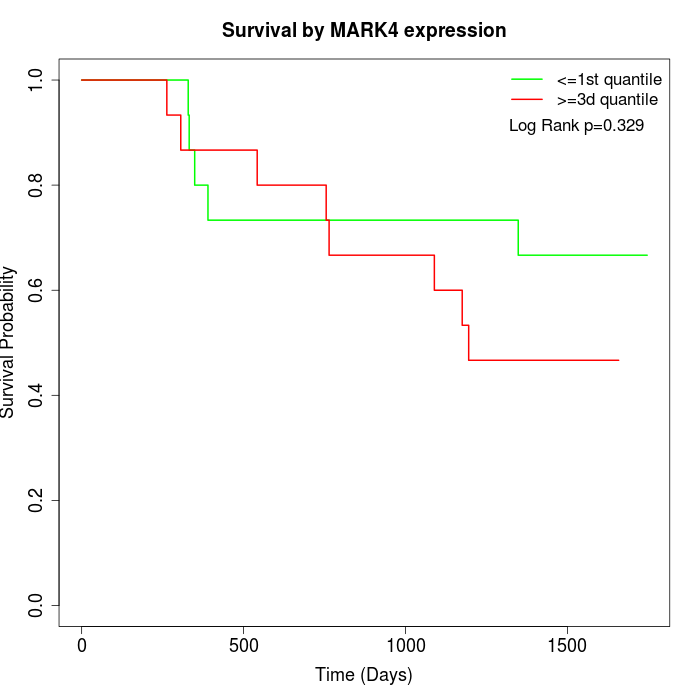
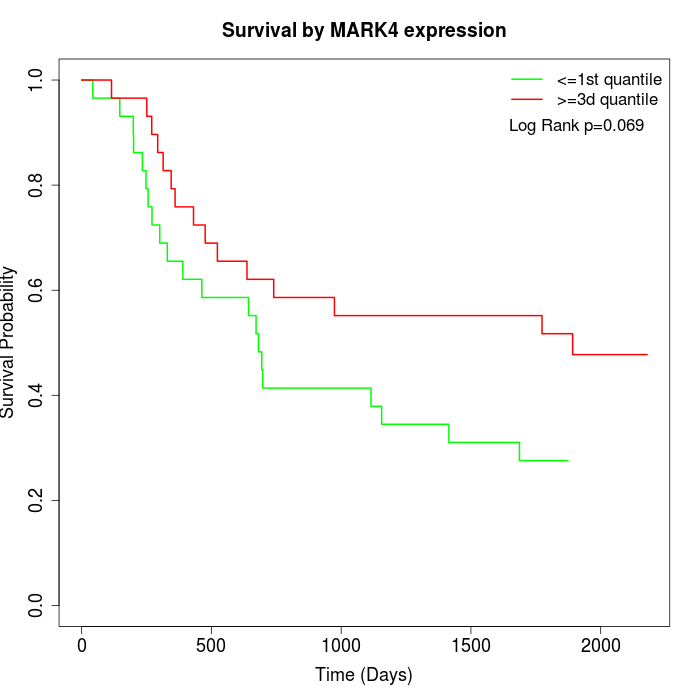
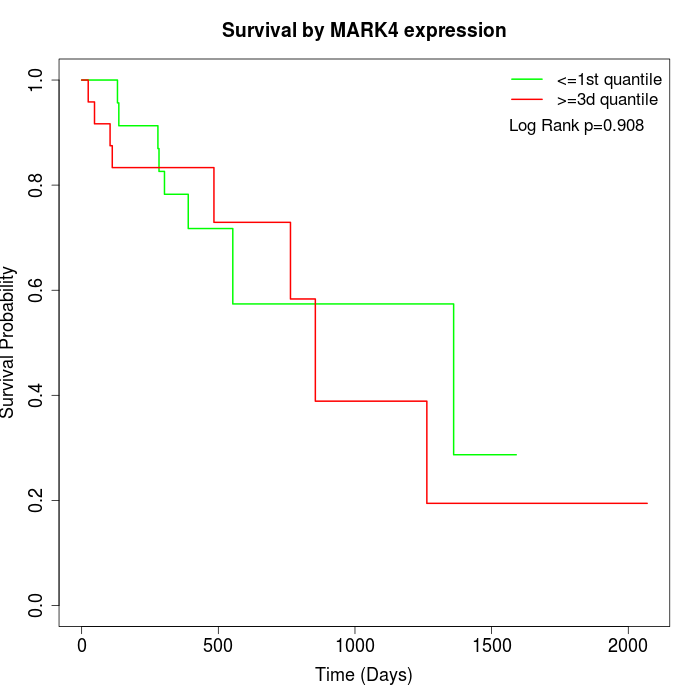
Survival by MARK4 expression:
Note: Click image to view full size file.
Copy number change of MARK4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MARK4 | 57787 | 4 | 5 | 21 | |
| GSE20123 | MARK4 | 57787 | 4 | 4 | 22 | |
| GSE43470 | MARK4 | 57787 | 3 | 11 | 29 | |
| GSE46452 | MARK4 | 57787 | 45 | 1 | 13 | |
| GSE47630 | MARK4 | 57787 | 8 | 6 | 26 | |
| GSE54993 | MARK4 | 57787 | 17 | 4 | 49 | |
| GSE54994 | MARK4 | 57787 | 6 | 12 | 35 | |
| GSE60625 | MARK4 | 57787 | 9 | 0 | 2 | |
| GSE74703 | MARK4 | 57787 | 3 | 7 | 26 | |
| GSE74704 | MARK4 | 57787 | 4 | 2 | 14 | |
| TCGA | MARK4 | 57787 | 14 | 18 | 64 |
Total number of gains: 117; Total number of losses: 70; Total Number of normals: 301.
Somatic mutations of MARK4:
Generating mutation plots.
Highly correlated genes for MARK4:
Showing top 20/428 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MARK4 | CASC15 | 0.785778 | 3 | 0 | 3 |
| MARK4 | GAK | 0.76604 | 3 | 0 | 3 |
| MARK4 | KRTAP9-6 | 0.746882 | 3 | 0 | 3 |
| MARK4 | GPR137C | 0.745368 | 3 | 0 | 3 |
| MARK4 | MIRLET7BHG | 0.742326 | 3 | 0 | 3 |
| MARK4 | NAPSA | 0.738871 | 3 | 0 | 3 |
| MARK4 | ERICH4 | 0.7338 | 3 | 0 | 3 |
| MARK4 | CLDN19 | 0.731986 | 3 | 0 | 3 |
| MARK4 | NOP58 | 0.725522 | 3 | 0 | 3 |
| MARK4 | KIR3DL2 | 0.725032 | 3 | 0 | 3 |
| MARK4 | IGF2 | 0.719864 | 3 | 0 | 3 |
| MARK4 | CCAR1 | 0.719528 | 3 | 0 | 3 |
| MARK4 | EVI5L | 0.717314 | 3 | 0 | 3 |
| MARK4 | PTPN23 | 0.7087 | 3 | 0 | 3 |
| MARK4 | GRINA | 0.708005 | 4 | 0 | 3 |
| MARK4 | KHSRP | 0.707995 | 6 | 0 | 6 |
| MARK4 | ZNF316 | 0.705618 | 5 | 0 | 4 |
| MARK4 | ALKBH6 | 0.703123 | 4 | 0 | 3 |
| MARK4 | LMLN | 0.701845 | 5 | 0 | 5 |
| MARK4 | HOXD13 | 0.701654 | 4 | 0 | 3 |
For details and further investigation, click here