| Full name: insulin like growth factor 2 | Alias Symbol: FLJ44734|IGF-II | ||
| Type: protein-coding gene | Cytoband: 11p15.5 | ||
| Entrez ID: 3481 | HGNC ID: HGNC:5466 | Ensembl Gene: ENSG00000167244 | OMIM ID: 147470 |
| Drug and gene relationship at DGIdb | |||
IGF2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05205 | Proteoglycans in cancer |
Expression of IGF2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | IGF2 | 3481 | 22901 | -0.1344 | 0.0101 | |
| GSE53624 | IGF2 | 3481 | 22901 | -0.0846 | 0.1114 | |
| GSE97050 | IGF2 | 3481 | A_23_P421379 | 0.6562 | 0.3909 | |
| SRP007169 | IGF2 | 3481 | RNAseq | 3.4725 | 0.0000 | |
| SRP008496 | IGF2 | 3481 | RNAseq | 4.5168 | 0.0000 | |
| SRP064894 | IGF2 | 3481 | RNAseq | 0.0637 | 0.8752 | |
| SRP133303 | IGF2 | 3481 | RNAseq | 0.8552 | 0.0476 | |
| SRP159526 | IGF2 | 3481 | RNAseq | 0.0936 | 0.8470 | |
| SRP193095 | IGF2 | 3481 | RNAseq | 3.2835 | 0.0000 | |
| SRP219564 | IGF2 | 3481 | RNAseq | 0.8687 | 0.2623 | |
| TCGA | IGF2 | 3481 | RNAseq | 0.0835 | 0.5531 |
Upregulated datasets: 3; Downregulated datasets: 0.
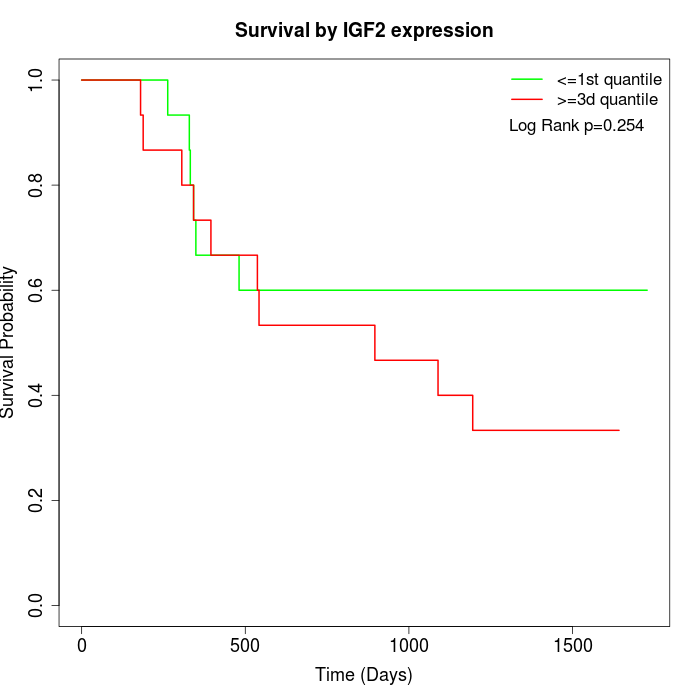
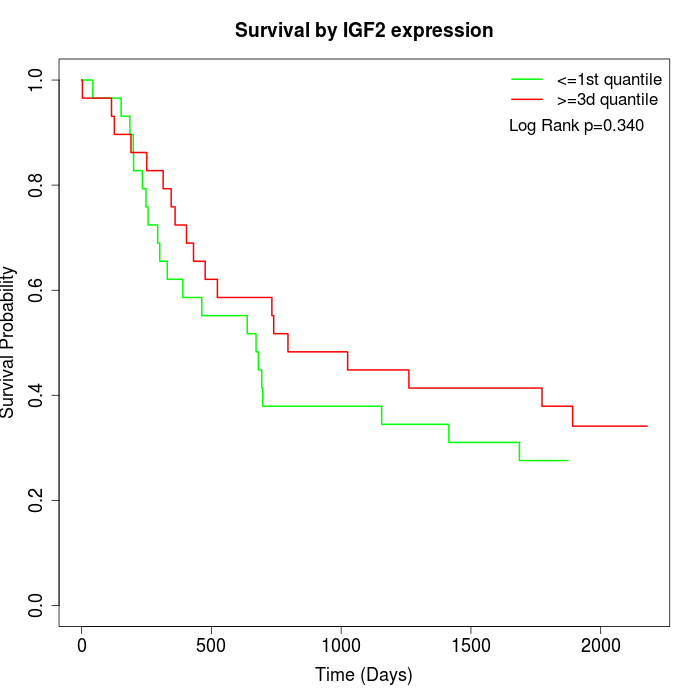
Survival by IGF2 expression:
Note: Click image to view full size file.
Copy number change of IGF2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | IGF2 | 3481 | 1 | 11 | 18 | |
| GSE20123 | IGF2 | 3481 | 1 | 12 | 17 | |
| GSE43470 | IGF2 | 3481 | 1 | 9 | 33 | |
| GSE46452 | IGF2 | 3481 | 7 | 8 | 44 | |
| GSE47630 | IGF2 | 3481 | 4 | 12 | 24 | |
| GSE54993 | IGF2 | 3481 | 3 | 1 | 66 | |
| GSE54994 | IGF2 | 3481 | 1 | 12 | 40 | |
| GSE60625 | IGF2 | 3481 | 0 | 0 | 11 | |
| GSE74703 | IGF2 | 3481 | 1 | 7 | 28 | |
| GSE74704 | IGF2 | 3481 | 0 | 8 | 12 | |
| TCGA | IGF2 | 3481 | 8 | 39 | 49 |
Total number of gains: 27; Total number of losses: 119; Total Number of normals: 342.
Somatic mutations of IGF2:
Generating mutation plots.
Highly correlated genes for IGF2:
Showing top 20/26 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| IGF2 | IFFO1 | 0.77231 | 3 | 0 | 3 |
| IGF2 | ADAMTSL2 | 0.720979 | 3 | 0 | 3 |
| IGF2 | MARK4 | 0.719864 | 3 | 0 | 3 |
| IGF2 | IMPDH1 | 0.696949 | 3 | 0 | 3 |
| IGF2 | RSPH9 | 0.679933 | 3 | 0 | 3 |
| IGF2 | FLT4 | 0.677025 | 3 | 0 | 3 |
| IGF2 | CBFA2T3 | 0.664555 | 3 | 0 | 3 |
| IGF2 | CCNB3 | 0.656417 | 3 | 0 | 3 |
| IGF2 | CARD11 | 0.645721 | 3 | 0 | 3 |
| IGF2 | LENG8 | 0.642231 | 3 | 0 | 3 |
| IGF2 | HES6 | 0.642002 | 3 | 0 | 3 |
| IGF2 | TFEB | 0.641544 | 3 | 0 | 3 |
| IGF2 | ZNF132 | 0.63404 | 3 | 0 | 3 |
| IGF2 | C16orf86 | 0.611975 | 3 | 0 | 3 |
| IGF2 | SOX17 | 0.608027 | 3 | 0 | 3 |
| IGF2 | DUSP8 | 0.605899 | 3 | 0 | 3 |
| IGF2 | PLEKHG5 | 0.604778 | 3 | 0 | 3 |
| IGF2 | OMP | 0.604216 | 3 | 0 | 3 |
| IGF2 | UTF1 | 0.594069 | 3 | 0 | 3 |
| IGF2 | CNTNAP1 | 0.592354 | 3 | 0 | 3 |
For details and further investigation, click here