| Full name: napsin A aspartic peptidase | Alias Symbol: NAP1|NAPA|Kdap|KAP | ||
| Type: protein-coding gene | Cytoband: 19q13.33 | ||
| Entrez ID: 9476 | HGNC ID: HGNC:13395 | Ensembl Gene: ENSG00000131400 | OMIM ID: 605631 |
| Drug and gene relationship at DGIdb | |||
Expression of NAPSA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NAPSA | 9476 | 223806_s_at | 0.0189 | 0.9701 | |
| GSE26886 | NAPSA | 9476 | 223806_s_at | 0.3117 | 0.0128 | |
| GSE45670 | NAPSA | 9476 | 223806_s_at | -0.0056 | 0.9609 | |
| GSE53622 | NAPSA | 9476 | 137631 | -0.3939 | 0.0000 | |
| GSE53624 | NAPSA | 9476 | 137631 | -0.6761 | 0.0000 | |
| GSE63941 | NAPSA | 9476 | 223806_s_at | 0.4041 | 0.0005 | |
| GSE77861 | NAPSA | 9476 | 223806_s_at | 0.0977 | 0.5071 | |
| GSE97050 | NAPSA | 9476 | A_32_P107029 | 0.6366 | 0.2233 | |
| SRP133303 | NAPSA | 9476 | RNAseq | 0.6559 | 0.0569 | |
| SRP159526 | NAPSA | 9476 | RNAseq | 0.4090 | 0.4114 | |
| SRP219564 | NAPSA | 9476 | RNAseq | 0.1319 | 0.8159 | |
| TCGA | NAPSA | 9476 | RNAseq | 0.7367 | 0.1583 |
Upregulated datasets: 0; Downregulated datasets: 0.
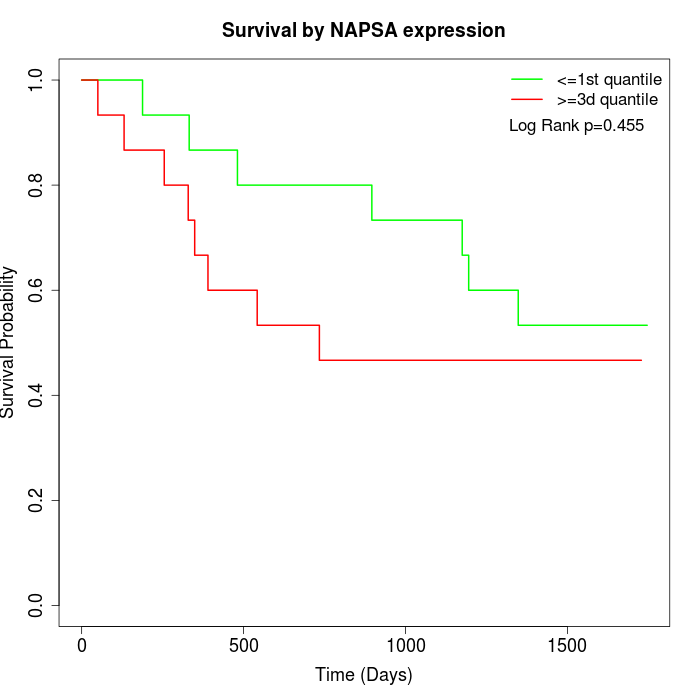
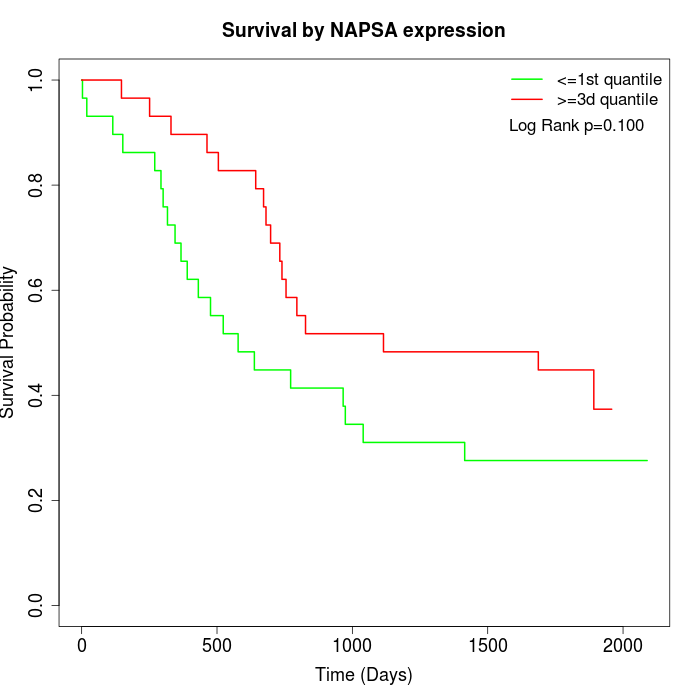
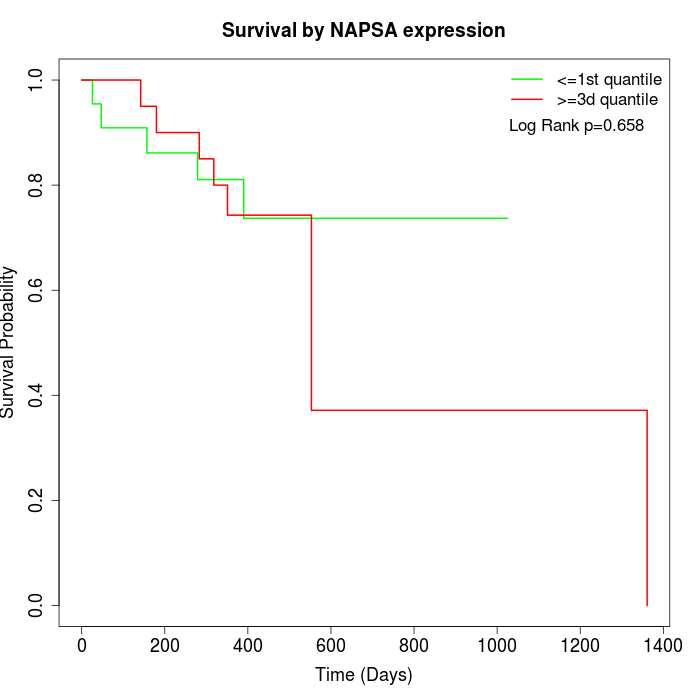
Survival by NAPSA expression:
Note: Click image to view full size file.
Copy number change of NAPSA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NAPSA | 9476 | 4 | 4 | 22 | |
| GSE20123 | NAPSA | 9476 | 4 | 3 | 23 | |
| GSE43470 | NAPSA | 9476 | 4 | 12 | 27 | |
| GSE46452 | NAPSA | 9476 | 45 | 1 | 13 | |
| GSE47630 | NAPSA | 9476 | 10 | 6 | 24 | |
| GSE54993 | NAPSA | 9476 | 17 | 4 | 49 | |
| GSE54994 | NAPSA | 9476 | 4 | 14 | 35 | |
| GSE60625 | NAPSA | 9476 | 9 | 0 | 2 | |
| GSE74703 | NAPSA | 9476 | 4 | 8 | 24 | |
| GSE74704 | NAPSA | 9476 | 4 | 1 | 15 | |
| TCGA | NAPSA | 9476 | 14 | 19 | 63 |
Total number of gains: 119; Total number of losses: 72; Total Number of normals: 297.
Somatic mutations of NAPSA:
Generating mutation plots.
Highly correlated genes for NAPSA:
Showing top 20/148 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NAPSA | MARK4 | 0.738871 | 3 | 0 | 3 |
| NAPSA | KPTN | 0.721961 | 3 | 0 | 3 |
| NAPSA | PAFAH1B3 | 0.703729 | 3 | 0 | 3 |
| NAPSA | SNX8 | 0.699469 | 3 | 0 | 3 |
| NAPSA | KLHDC7B | 0.681592 | 4 | 0 | 3 |
| NAPSA | PCSK1N | 0.680982 | 3 | 0 | 3 |
| NAPSA | TNK2 | 0.674564 | 3 | 0 | 3 |
| NAPSA | CD52 | 0.673857 | 5 | 0 | 5 |
| NAPSA | RAC3 | 0.654945 | 4 | 0 | 4 |
| NAPSA | FGFR4 | 0.654062 | 4 | 0 | 3 |
| NAPSA | MCM5 | 0.65114 | 4 | 0 | 3 |
| NAPSA | RIN1 | 0.643773 | 3 | 0 | 3 |
| NAPSA | MICALL2 | 0.643708 | 4 | 0 | 3 |
| NAPSA | EEFSEC | 0.637766 | 3 | 0 | 3 |
| NAPSA | PLD4 | 0.637378 | 5 | 0 | 5 |
| NAPSA | RUVBL2 | 0.636225 | 5 | 0 | 4 |
| NAPSA | FURIN | 0.634215 | 3 | 0 | 3 |
| NAPSA | FCRLA | 0.633705 | 3 | 0 | 3 |
| NAPSA | CR2 | 0.633093 | 4 | 0 | 3 |
| NAPSA | PAX5 | 0.630434 | 5 | 0 | 4 |
For details and further investigation, click here