| Full name: megakaryocyte-associated tyrosine kinase | Alias Symbol: HYLTK|CTK|HYL|Lsk|CHK|HHYLTK|DKFZp434N1212|MGC1708|MGC2101 | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 4145 | HGNC ID: HGNC:6906 | Ensembl Gene: ENSG00000007264 | OMIM ID: 600038 |
| Drug and gene relationship at DGIdb | |||
MATK involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04722 | Neurotrophin signaling pathway |
Expression of MATK:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MATK | 4145 | 206267_s_at | 0.1402 | 0.6955 | |
| GSE20347 | MATK | 4145 | 206267_s_at | 0.0002 | 0.9989 | |
| GSE23400 | MATK | 4145 | 206267_s_at | -0.1030 | 0.0016 | |
| GSE26886 | MATK | 4145 | 206267_s_at | -0.0667 | 0.6091 | |
| GSE29001 | MATK | 4145 | 206267_s_at | 0.0603 | 0.8232 | |
| GSE38129 | MATK | 4145 | 206267_s_at | 0.0755 | 0.5143 | |
| GSE45670 | MATK | 4145 | 206267_s_at | -0.0180 | 0.8915 | |
| GSE53622 | MATK | 4145 | 48169 | -0.0266 | 0.7270 | |
| GSE53624 | MATK | 4145 | 48169 | 0.1328 | 0.1609 | |
| GSE63941 | MATK | 4145 | 206267_s_at | 0.3732 | 0.2791 | |
| GSE77861 | MATK | 4145 | 206267_s_at | 0.0697 | 0.6642 | |
| GSE97050 | MATK | 4145 | A_23_P50678 | -0.1257 | 0.5698 | |
| SRP064894 | MATK | 4145 | RNAseq | 0.5648 | 0.1534 | |
| SRP133303 | MATK | 4145 | RNAseq | -0.5038 | 0.0524 | |
| SRP159526 | MATK | 4145 | RNAseq | -0.4933 | 0.4102 | |
| TCGA | MATK | 4145 | RNAseq | 0.9436 | 0.0049 |
Upregulated datasets: 0; Downregulated datasets: 0.
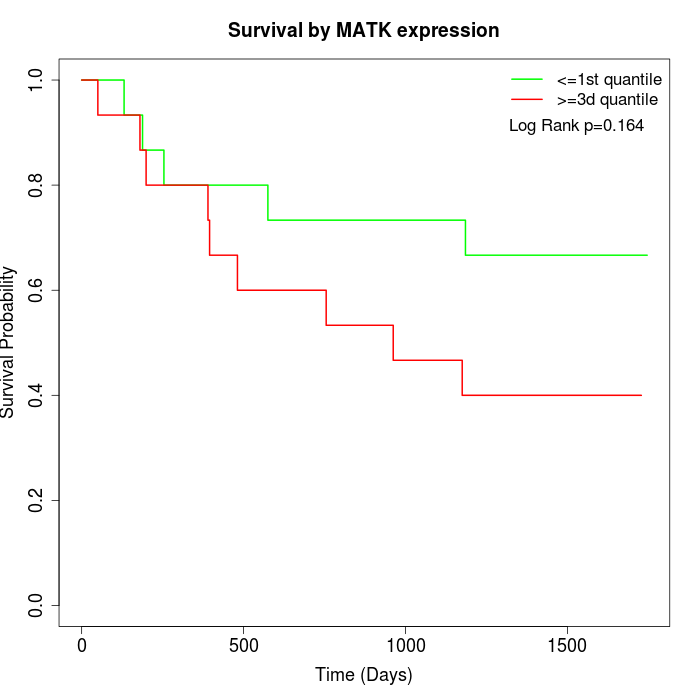
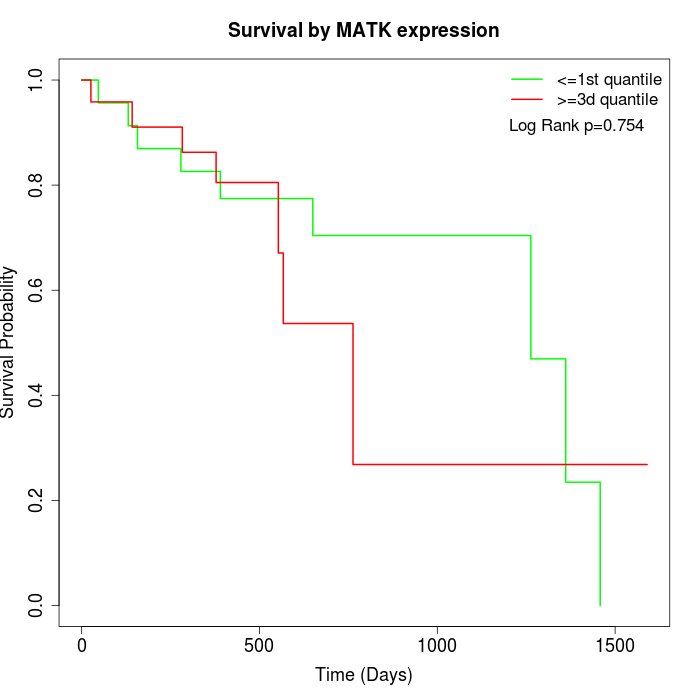
Survival by MATK expression:
Note: Click image to view full size file.
Copy number change of MATK:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MATK | 4145 | 5 | 3 | 22 | |
| GSE20123 | MATK | 4145 | 4 | 2 | 24 | |
| GSE43470 | MATK | 4145 | 1 | 9 | 33 | |
| GSE46452 | MATK | 4145 | 47 | 1 | 11 | |
| GSE47630 | MATK | 4145 | 5 | 7 | 28 | |
| GSE54993 | MATK | 4145 | 16 | 3 | 51 | |
| GSE54994 | MATK | 4145 | 8 | 16 | 29 | |
| GSE60625 | MATK | 4145 | 9 | 0 | 2 | |
| GSE74703 | MATK | 4145 | 1 | 6 | 29 | |
| GSE74704 | MATK | 4145 | 1 | 2 | 17 | |
| TCGA | MATK | 4145 | 10 | 19 | 67 |
Total number of gains: 107; Total number of losses: 68; Total Number of normals: 313.
Somatic mutations of MATK:
Generating mutation plots.
Highly correlated genes for MATK:
Showing top 20/130 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MATK | SBNO2 | 0.727762 | 3 | 0 | 3 |
| MATK | GNL1 | 0.716055 | 3 | 0 | 3 |
| MATK | CPN1 | 0.701913 | 3 | 0 | 3 |
| MATK | SCUBE1 | 0.699696 | 3 | 0 | 3 |
| MATK | SCGN | 0.684624 | 3 | 0 | 3 |
| MATK | NOXA1 | 0.682548 | 3 | 0 | 3 |
| MATK | SALL3 | 0.679302 | 3 | 0 | 3 |
| MATK | RPIA | 0.676393 | 3 | 0 | 3 |
| MATK | OSBPL5 | 0.67432 | 3 | 0 | 3 |
| MATK | HOTAIR | 0.663208 | 4 | 0 | 3 |
| MATK | ATAD3C | 0.663036 | 4 | 0 | 3 |
| MATK | B3GAT3 | 0.660922 | 4 | 0 | 3 |
| MATK | FBXO24 | 0.660138 | 4 | 0 | 3 |
| MATK | PCP2 | 0.655167 | 3 | 0 | 3 |
| MATK | SLC26A1 | 0.650909 | 5 | 0 | 4 |
| MATK | LTB4R | 0.649686 | 4 | 0 | 4 |
| MATK | JSRP1 | 0.642275 | 5 | 0 | 3 |
| MATK | KIFC2 | 0.639272 | 4 | 0 | 3 |
| MATK | CPNE9 | 0.638649 | 5 | 0 | 3 |
| MATK | SLC25A47 | 0.634072 | 4 | 0 | 3 |
For details and further investigation, click here