| Full name: minichromosome maintenance 9 homologous recombination repair factor | Alias Symbol: MGC35304|dJ329L24.3|FLJ20170 | ||
| Type: protein-coding gene | Cytoband: 6q22.31 | ||
| Entrez ID: 254394 | HGNC ID: HGNC:21484 | Ensembl Gene: ENSG00000111877 | OMIM ID: 610098 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of MCM9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MCM9 | 254394 | 219673_at | 0.0535 | 0.9599 | |
| GSE20347 | MCM9 | 254394 | 219673_at | -0.1777 | 0.3733 | |
| GSE23400 | MCM9 | 254394 | 219673_at | 0.1346 | 0.0050 | |
| GSE26886 | MCM9 | 254394 | 219673_at | -0.3848 | 0.1701 | |
| GSE29001 | MCM9 | 254394 | 219673_at | 0.1448 | 0.6883 | |
| GSE38129 | MCM9 | 254394 | 219673_at | 0.1033 | 0.5202 | |
| GSE45670 | MCM9 | 254394 | 219673_at | -0.0033 | 0.9867 | |
| GSE53622 | MCM9 | 254394 | 80486 | 0.0419 | 0.7274 | |
| GSE53624 | MCM9 | 254394 | 80486 | 0.1145 | 0.3040 | |
| GSE63941 | MCM9 | 254394 | 219673_at | 0.0296 | 0.9624 | |
| GSE77861 | MCM9 | 254394 | 219673_at | 0.2546 | 0.5572 | |
| GSE97050 | MCM9 | 254394 | A_33_P3254555 | 0.3310 | 0.6504 | |
| SRP007169 | MCM9 | 254394 | RNAseq | 1.0465 | 0.0138 | |
| SRP008496 | MCM9 | 254394 | RNAseq | 1.3650 | 0.0001 | |
| SRP064894 | MCM9 | 254394 | RNAseq | 0.1623 | 0.4976 | |
| SRP133303 | MCM9 | 254394 | RNAseq | 0.0705 | 0.5494 | |
| SRP159526 | MCM9 | 254394 | RNAseq | 0.1570 | 0.4998 | |
| SRP193095 | MCM9 | 254394 | RNAseq | -0.1094 | 0.1768 | |
| SRP219564 | MCM9 | 254394 | RNAseq | 0.1388 | 0.7189 | |
| TCGA | MCM9 | 254394 | RNAseq | 0.1195 | 0.1328 |
Upregulated datasets: 2; Downregulated datasets: 0.
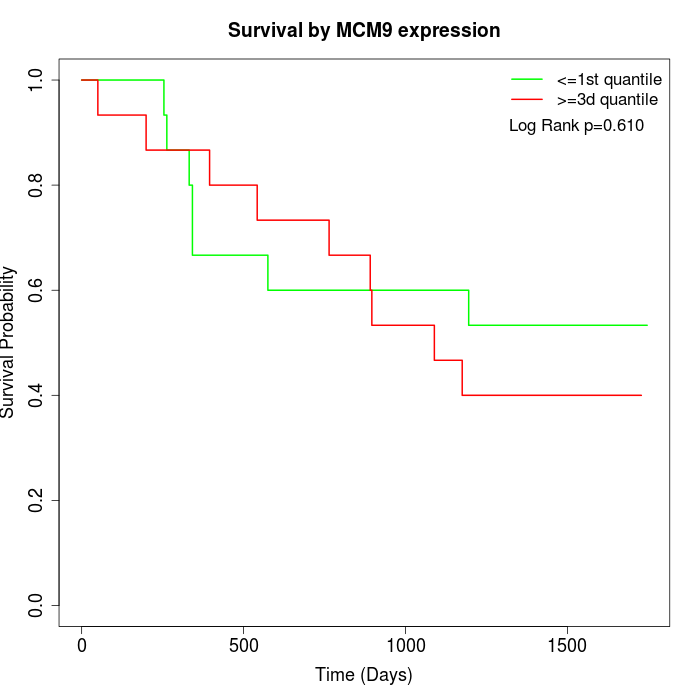
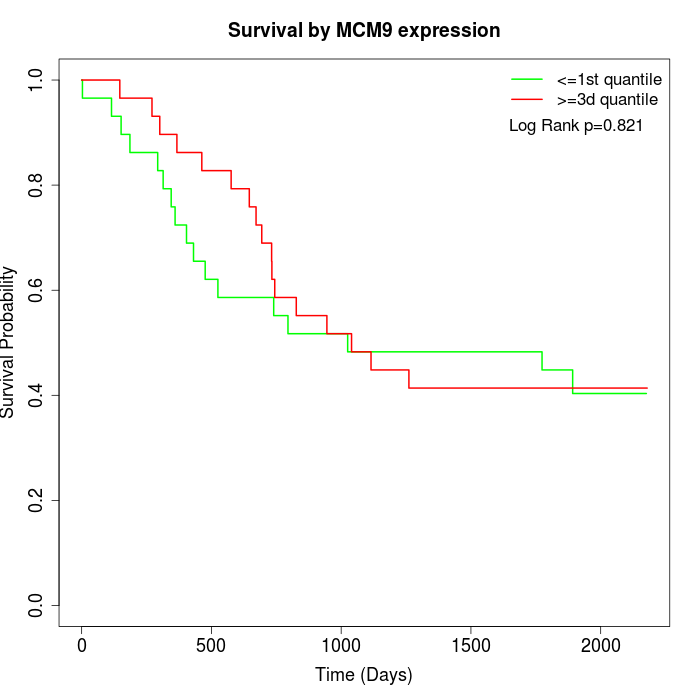
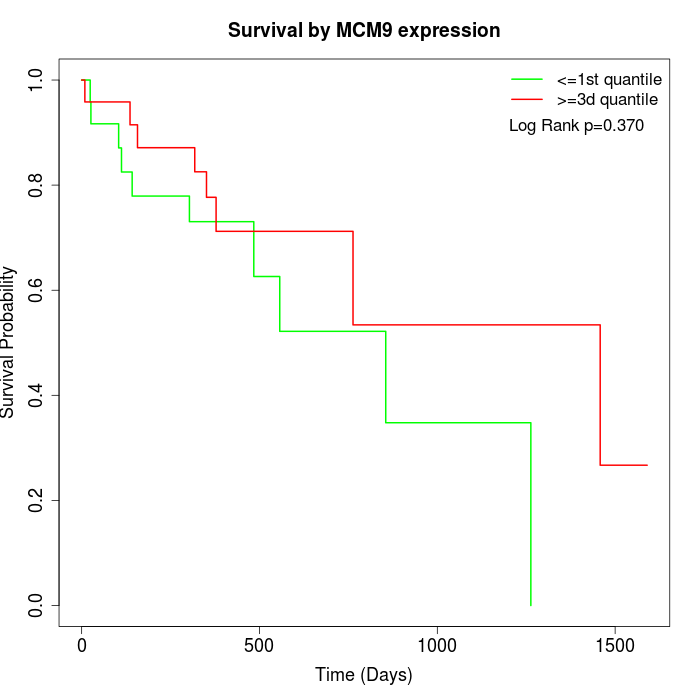
Survival by MCM9 expression:
Note: Click image to view full size file.
Copy number change of MCM9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MCM9 | 254394 | 1 | 4 | 25 | |
| GSE20123 | MCM9 | 254394 | 1 | 3 | 26 | |
| GSE43470 | MCM9 | 254394 | 4 | 2 | 37 | |
| GSE46452 | MCM9 | 254394 | 2 | 11 | 46 | |
| GSE47630 | MCM9 | 254394 | 9 | 4 | 27 | |
| GSE54993 | MCM9 | 254394 | 3 | 2 | 65 | |
| GSE54994 | MCM9 | 254394 | 8 | 8 | 37 | |
| GSE60625 | MCM9 | 254394 | 0 | 1 | 10 | |
| GSE74703 | MCM9 | 254394 | 3 | 2 | 31 | |
| GSE74704 | MCM9 | 254394 | 0 | 1 | 19 | |
| TCGA | MCM9 | 254394 | 10 | 21 | 65 |
Total number of gains: 41; Total number of losses: 59; Total Number of normals: 388.
Somatic mutations of MCM9:
Generating mutation plots.
Highly correlated genes for MCM9:
Showing top 20/45 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MCM9 | ZNF454 | 0.836967 | 3 | 0 | 3 |
| MCM9 | DCAF8L2 | 0.780227 | 3 | 0 | 3 |
| MCM9 | SERPINA11 | 0.774686 | 3 | 0 | 3 |
| MCM9 | ZSCAN5A | 0.692755 | 3 | 0 | 3 |
| MCM9 | GCH1 | 0.66631 | 3 | 0 | 3 |
| MCM9 | ZNF621 | 0.665139 | 6 | 0 | 5 |
| MCM9 | MRPL23 | 0.655183 | 3 | 0 | 3 |
| MCM9 | RTKN2 | 0.638205 | 3 | 0 | 3 |
| MCM9 | TFB1M | 0.630508 | 3 | 0 | 3 |
| MCM9 | DDIAS | 0.616323 | 3 | 0 | 3 |
| MCM9 | ATP6V0A2 | 0.611679 | 3 | 0 | 3 |
| MCM9 | GEMIN2 | 0.609211 | 3 | 0 | 3 |
| MCM9 | BAZ1A | 0.606416 | 3 | 0 | 3 |
| MCM9 | HS6ST2 | 0.601911 | 3 | 0 | 3 |
| MCM9 | CHAC2 | 0.601736 | 4 | 0 | 3 |
| MCM9 | ACAT2 | 0.59838 | 4 | 0 | 3 |
| MCM9 | PSMD6 | 0.581702 | 6 | 0 | 4 |
| MCM9 | GNAL | 0.572361 | 4 | 0 | 3 |
| MCM9 | PAXIP1 | 0.567997 | 7 | 0 | 4 |
| MCM9 | PHF10 | 0.559988 | 6 | 0 | 3 |
For details and further investigation, click here