| Full name: ChaC cation transport regulator homolog 2 | Alias Symbol: GCG1 | ||
| Type: protein-coding gene | Cytoband: 2p16.2 | ||
| Entrez ID: 494143 | HGNC ID: HGNC:32363 | Ensembl Gene: ENSG00000143942 | OMIM ID: 617446 |
| Drug and gene relationship at DGIdb | |||
Expression of CHAC2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CHAC2 | 494143 | 235117_at | 0.3483 | 0.7425 | |
| GSE26886 | CHAC2 | 494143 | 235117_at | -0.7675 | 0.0299 | |
| GSE45670 | CHAC2 | 494143 | 235117_at | 0.4621 | 0.0931 | |
| GSE63941 | CHAC2 | 494143 | 235117_at | 2.1474 | 0.0058 | |
| GSE77861 | CHAC2 | 494143 | 235117_at | 0.0156 | 0.9789 | |
| GSE97050 | CHAC2 | 494143 | A_32_P194264 | 0.4896 | 0.3867 | |
| SRP007169 | CHAC2 | 494143 | RNAseq | -0.0738 | 0.9045 | |
| SRP008496 | CHAC2 | 494143 | RNAseq | 0.0222 | 0.9613 | |
| SRP064894 | CHAC2 | 494143 | RNAseq | -0.0821 | 0.8013 | |
| SRP133303 | CHAC2 | 494143 | RNAseq | 0.7199 | 0.0393 | |
| SRP159526 | CHAC2 | 494143 | RNAseq | 1.3190 | 0.0293 | |
| SRP193095 | CHAC2 | 494143 | RNAseq | -0.0844 | 0.5588 | |
| SRP219564 | CHAC2 | 494143 | RNAseq | -0.0524 | 0.9398 | |
| TCGA | CHAC2 | 494143 | RNAseq | 0.2857 | 0.0126 |
Upregulated datasets: 2; Downregulated datasets: 0.
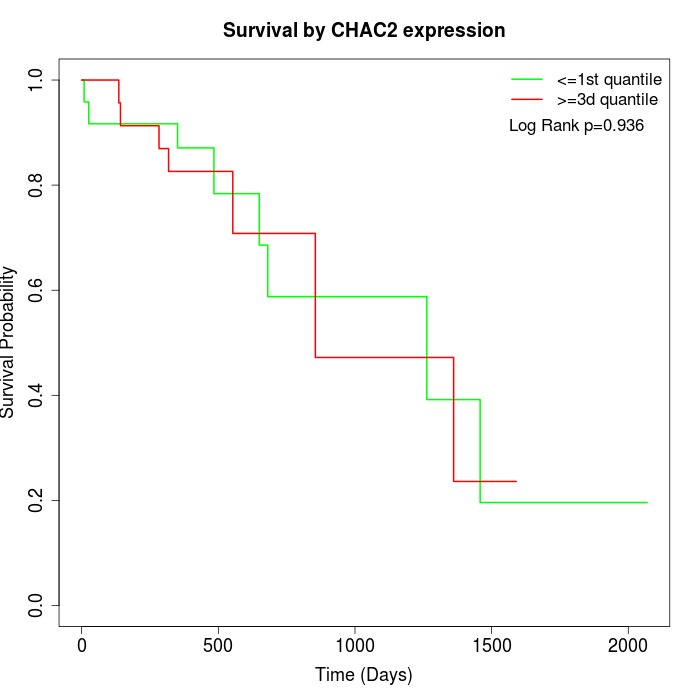
Survival by CHAC2 expression:
Note: Click image to view full size file.
Copy number change of CHAC2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CHAC2 | 494143 | 8 | 0 | 22 | |
| GSE20123 | CHAC2 | 494143 | 7 | 0 | 23 | |
| GSE43470 | CHAC2 | 494143 | 5 | 0 | 38 | |
| GSE46452 | CHAC2 | 494143 | 2 | 4 | 53 | |
| GSE47630 | CHAC2 | 494143 | 8 | 0 | 32 | |
| GSE54993 | CHAC2 | 494143 | 0 | 5 | 65 | |
| GSE54994 | CHAC2 | 494143 | 12 | 0 | 41 | |
| GSE60625 | CHAC2 | 494143 | 0 | 3 | 8 | |
| GSE74703 | CHAC2 | 494143 | 5 | 0 | 31 | |
| GSE74704 | CHAC2 | 494143 | 8 | 0 | 12 | |
| TCGA | CHAC2 | 494143 | 36 | 2 | 58 |
Total number of gains: 91; Total number of losses: 14; Total Number of normals: 383.
Somatic mutations of CHAC2:
Generating mutation plots.
Highly correlated genes for CHAC2:
Showing top 20/137 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CHAC2 | CPSF2 | 0.692606 | 3 | 0 | 3 |
| CHAC2 | CEP76 | 0.68544 | 3 | 0 | 3 |
| CHAC2 | MTFP1 | 0.676957 | 4 | 0 | 4 |
| CHAC2 | NCAPH | 0.672785 | 3 | 0 | 3 |
| CHAC2 | FLAD1 | 0.66222 | 3 | 0 | 3 |
| CHAC2 | YIF1B | 0.655644 | 3 | 0 | 3 |
| CHAC2 | MAP3K9 | 0.649686 | 3 | 0 | 3 |
| CHAC2 | FOXE1 | 0.64549 | 3 | 0 | 3 |
| CHAC2 | PNO1 | 0.645356 | 4 | 0 | 3 |
| CHAC2 | ALG8 | 0.642099 | 3 | 0 | 3 |
| CHAC2 | MREG | 0.641344 | 3 | 0 | 3 |
| CHAC2 | EPCAM | 0.639591 | 4 | 0 | 4 |
| CHAC2 | PC | 0.634296 | 3 | 0 | 3 |
| CHAC2 | ORC6 | 0.632299 | 4 | 0 | 4 |
| CHAC2 | TSPAN13 | 0.630161 | 4 | 0 | 3 |
| CHAC2 | FEN1 | 0.61973 | 3 | 0 | 3 |
| CHAC2 | MCM7 | 0.617381 | 3 | 0 | 3 |
| CHAC2 | CYB5R2 | 0.615972 | 4 | 0 | 3 |
| CHAC2 | PLK4 | 0.615842 | 5 | 0 | 3 |
| CHAC2 | SORD | 0.615748 | 4 | 0 | 3 |
For details and further investigation, click here