| Full name: mediator complex subunit 16 | Alias Symbol: DRIP92|TRAP95 | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 10025 | HGNC ID: HGNC:17556 | Ensembl Gene: ENSG00000175221 | OMIM ID: 604062 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of MED16:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MED16 | 10025 | 43544_at | 0.0848 | 0.8585 | |
| GSE20347 | MED16 | 10025 | 43544_at | -0.4665 | 0.0009 | |
| GSE23400 | MED16 | 10025 | 221938_x_at | -0.2667 | 0.0000 | |
| GSE26886 | MED16 | 10025 | 43544_at | 0.4744 | 0.0919 | |
| GSE29001 | MED16 | 10025 | 43544_at | -0.3117 | 0.2092 | |
| GSE38129 | MED16 | 10025 | 43544_at | -0.2573 | 0.0320 | |
| GSE45670 | MED16 | 10025 | 43544_at | 0.1228 | 0.4190 | |
| GSE53622 | MED16 | 10025 | 76481 | -0.2994 | 0.0000 | |
| GSE53624 | MED16 | 10025 | 47682 | -2.0545 | 0.0000 | |
| GSE63941 | MED16 | 10025 | 43544_at | -1.1775 | 0.0078 | |
| GSE77861 | MED16 | 10025 | 221545_x_at | -0.1587 | 0.3816 | |
| GSE97050 | MED16 | 10025 | A_23_P56170 | 0.3422 | 0.2413 | |
| SRP007169 | MED16 | 10025 | RNAseq | -1.3087 | 0.0006 | |
| SRP008496 | MED16 | 10025 | RNAseq | -1.3984 | 0.0000 | |
| SRP064894 | MED16 | 10025 | RNAseq | 0.2781 | 0.3647 | |
| SRP133303 | MED16 | 10025 | RNAseq | -0.1435 | 0.3971 | |
| SRP159526 | MED16 | 10025 | RNAseq | -0.3054 | 0.1118 | |
| SRP193095 | MED16 | 10025 | RNAseq | -0.3562 | 0.0295 | |
| SRP219564 | MED16 | 10025 | RNAseq | 0.0541 | 0.8106 | |
| TCGA | MED16 | 10025 | RNAseq | -0.0430 | 0.3585 |
Upregulated datasets: 0; Downregulated datasets: 4.
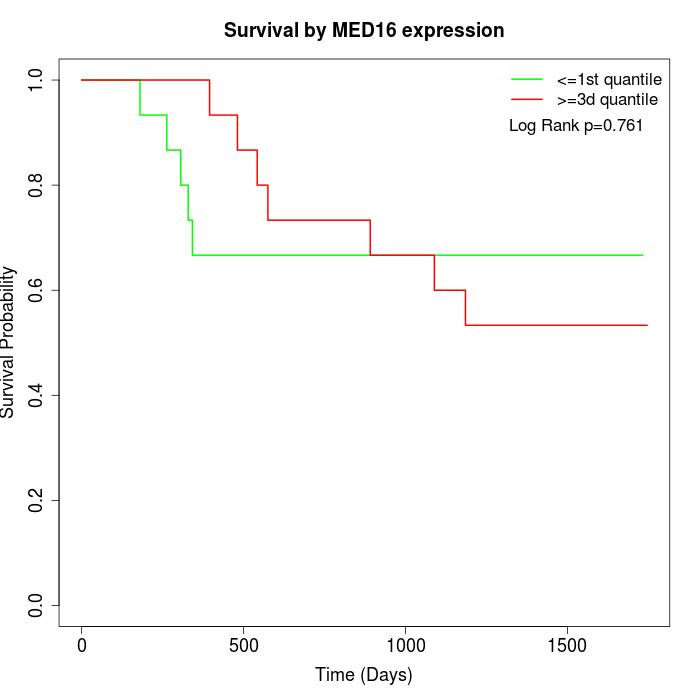
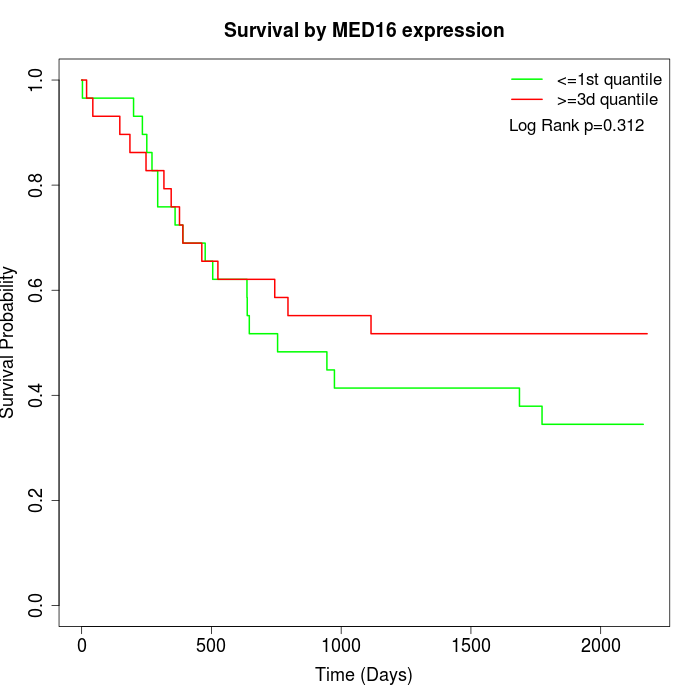
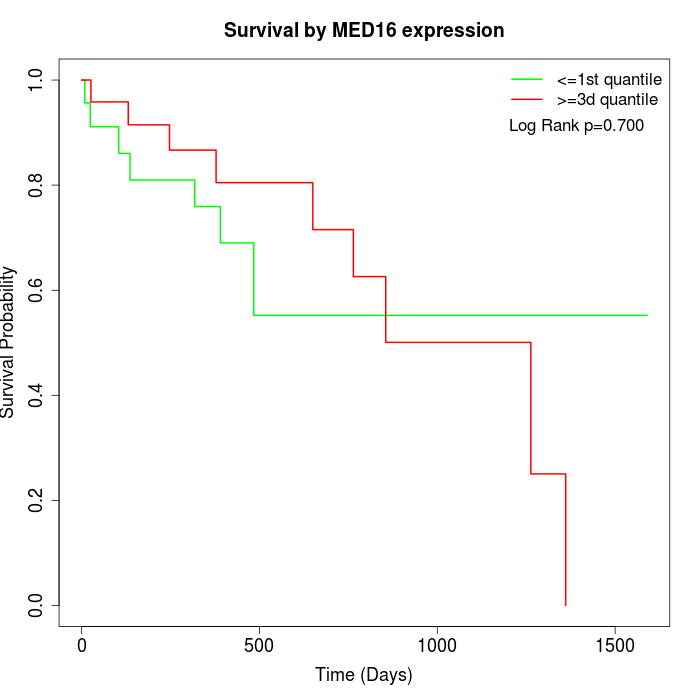
Survival by MED16 expression:
Note: Click image to view full size file.
Copy number change of MED16:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MED16 | 10025 | 4 | 6 | 20 | |
| GSE20123 | MED16 | 10025 | 4 | 5 | 21 | |
| GSE43470 | MED16 | 10025 | 1 | 9 | 33 | |
| GSE46452 | MED16 | 10025 | 47 | 1 | 11 | |
| GSE47630 | MED16 | 10025 | 5 | 7 | 28 | |
| GSE54993 | MED16 | 10025 | 13 | 4 | 53 | |
| GSE54994 | MED16 | 10025 | 8 | 15 | 30 | |
| GSE60625 | MED16 | 10025 | 9 | 0 | 2 | |
| GSE74703 | MED16 | 10025 | 1 | 7 | 28 | |
| GSE74704 | MED16 | 10025 | 3 | 3 | 14 | |
| TCGA | MED16 | 10025 | 8 | 25 | 63 |
Total number of gains: 103; Total number of losses: 82; Total Number of normals: 303.
Somatic mutations of MED16:
Generating mutation plots.
Highly correlated genes for MED16:
Showing top 20/160 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MED16 | BIN3 | 0.695984 | 3 | 0 | 3 |
| MED16 | HES5 | 0.646453 | 3 | 0 | 3 |
| MED16 | CPTP | 0.645744 | 6 | 0 | 6 |
| MED16 | KMT2D | 0.644424 | 3 | 0 | 3 |
| MED16 | MICALL2 | 0.643424 | 3 | 0 | 3 |
| MED16 | ABCC10 | 0.624683 | 4 | 0 | 4 |
| MED16 | SCFD2 | 0.612303 | 4 | 0 | 4 |
| MED16 | YPEL3 | 0.611848 | 3 | 0 | 3 |
| MED16 | ELOVL1 | 0.60406 | 3 | 0 | 3 |
| MED16 | CTDP1 | 0.602998 | 5 | 0 | 4 |
| MED16 | IL34 | 0.600228 | 4 | 0 | 3 |
| MED16 | TRIM55 | 0.588328 | 3 | 0 | 3 |
| MED16 | RPS6KB2 | 0.588043 | 8 | 0 | 7 |
| MED16 | ANO10 | 0.587862 | 7 | 0 | 6 |
| MED16 | RNPEPL1 | 0.587199 | 7 | 0 | 7 |
| MED16 | ZC3H3 | 0.579504 | 4 | 0 | 3 |
| MED16 | ARSF | 0.577287 | 6 | 0 | 5 |
| MED16 | CORO1B | 0.576152 | 8 | 0 | 5 |
| MED16 | MICAL1 | 0.576017 | 4 | 0 | 3 |
| MED16 | B4GALT7 | 0.57549 | 5 | 0 | 4 |
For details and further investigation, click here