| Full name: MICAL like 2 | Alias Symbol: MGC46023|FLJ23471|MICAL-L2|JRAB | ||
| Type: protein-coding gene | Cytoband: 7p22.3 | ||
| Entrez ID: 79778 | HGNC ID: HGNC:29672 | Ensembl Gene: ENSG00000164877 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of MICALL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MICALL2 | 79778 | 219332_at | 0.4194 | 0.3483 | |
| GSE20347 | MICALL2 | 79778 | 219332_at | 0.3976 | 0.0545 | |
| GSE23400 | MICALL2 | 79778 | 219332_at | 0.3040 | 0.0002 | |
| GSE26886 | MICALL2 | 79778 | 219332_at | 0.9942 | 0.0010 | |
| GSE29001 | MICALL2 | 79778 | 219332_at | 0.4201 | 0.1672 | |
| GSE38129 | MICALL2 | 79778 | 219332_at | 0.3632 | 0.0229 | |
| GSE45670 | MICALL2 | 79778 | 219332_at | 0.2897 | 0.2032 | |
| GSE53622 | MICALL2 | 79778 | 12948 | 0.2242 | 0.0035 | |
| GSE53624 | MICALL2 | 79778 | 12948 | 0.3133 | 0.0002 | |
| GSE63941 | MICALL2 | 79778 | 219332_at | -0.2396 | 0.6946 | |
| GSE77861 | MICALL2 | 79778 | 241478_at | -0.0653 | 0.7179 | |
| GSE97050 | MICALL2 | 79778 | A_24_P303524 | 0.7387 | 0.1314 | |
| SRP007169 | MICALL2 | 79778 | RNAseq | 1.0046 | 0.1290 | |
| SRP008496 | MICALL2 | 79778 | RNAseq | 1.4659 | 0.0027 | |
| SRP064894 | MICALL2 | 79778 | RNAseq | 0.8524 | 0.0010 | |
| SRP133303 | MICALL2 | 79778 | RNAseq | 0.3028 | 0.1585 | |
| SRP159526 | MICALL2 | 79778 | RNAseq | 0.1313 | 0.7049 | |
| SRP193095 | MICALL2 | 79778 | RNAseq | 0.5933 | 0.0186 | |
| SRP219564 | MICALL2 | 79778 | RNAseq | 0.6451 | 0.0284 | |
| TCGA | MICALL2 | 79778 | RNAseq | -0.0836 | 0.2644 |
Upregulated datasets: 1; Downregulated datasets: 0.
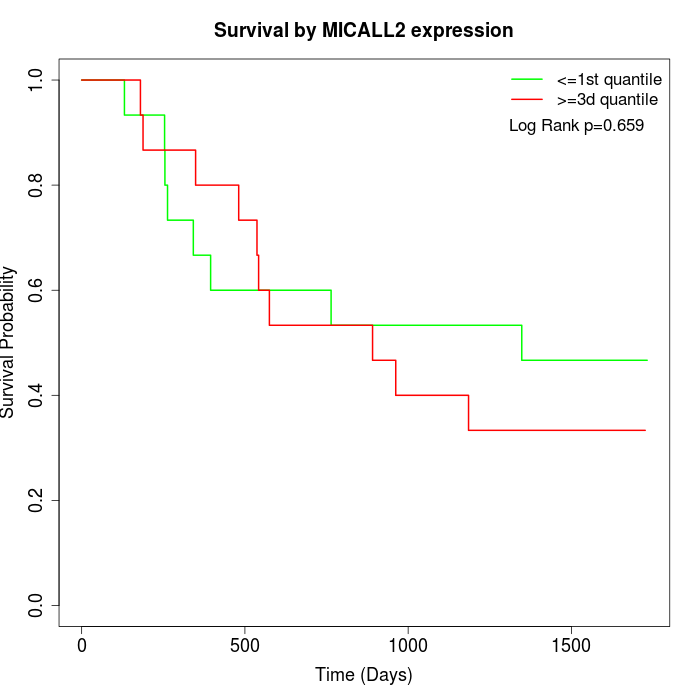
Survival by MICALL2 expression:
Note: Click image to view full size file.
Copy number change of MICALL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MICALL2 | 79778 | 14 | 0 | 16 | |
| GSE20123 | MICALL2 | 79778 | 14 | 0 | 16 | |
| GSE43470 | MICALL2 | 79778 | 6 | 2 | 35 | |
| GSE46452 | MICALL2 | 79778 | 13 | 1 | 45 | |
| GSE47630 | MICALL2 | 79778 | 10 | 1 | 29 | |
| GSE54993 | MICALL2 | 79778 | 0 | 7 | 63 | |
| GSE54994 | MICALL2 | 79778 | 19 | 2 | 32 | |
| GSE60625 | MICALL2 | 79778 | 0 | 0 | 11 | |
| GSE74703 | MICALL2 | 79778 | 6 | 1 | 29 | |
| GSE74704 | MICALL2 | 79778 | 9 | 0 | 11 | |
| TCGA | MICALL2 | 79778 | 57 | 5 | 34 |
Total number of gains: 148; Total number of losses: 19; Total Number of normals: 321.
Somatic mutations of MICALL2:
Generating mutation plots.
Highly correlated genes for MICALL2:
Showing top 20/251 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MICALL2 | ADAMTSL2 | 0.777085 | 3 | 0 | 3 |
| MICALL2 | LINGO1 | 0.749049 | 3 | 0 | 3 |
| MICALL2 | TMEM190 | 0.712016 | 3 | 0 | 3 |
| MICALL2 | ADCK5 | 0.702969 | 4 | 0 | 3 |
| MICALL2 | LTB4R2 | 0.701446 | 3 | 0 | 3 |
| MICALL2 | TMC8 | 0.694996 | 4 | 0 | 3 |
| MICALL2 | CHST13 | 0.682488 | 3 | 0 | 3 |
| MICALL2 | TIGD5 | 0.681717 | 3 | 0 | 3 |
| MICALL2 | MAST2 | 0.674647 | 3 | 0 | 3 |
| MICALL2 | ZNF574 | 0.668755 | 4 | 0 | 3 |
| MICALL2 | SHARPIN | 0.66736 | 4 | 0 | 4 |
| MICALL2 | ZNF683 | 0.657809 | 3 | 0 | 3 |
| MICALL2 | COL13A1 | 0.657358 | 3 | 0 | 3 |
| MICALL2 | ADTRP | 0.655332 | 3 | 0 | 3 |
| MICALL2 | C4orf48 | 0.650191 | 5 | 0 | 4 |
| MICALL2 | MYO1G | 0.64763 | 4 | 0 | 3 |
| MICALL2 | EPHB2 | 0.64756 | 5 | 0 | 3 |
| MICALL2 | ZDHHC18 | 0.645153 | 4 | 0 | 3 |
| MICALL2 | NAPSA | 0.643708 | 4 | 0 | 3 |
| MICALL2 | MED16 | 0.643424 | 3 | 0 | 3 |
For details and further investigation, click here