| Full name: tripartite motif containing 55 | Alias Symbol: MURF-2 | ||
| Type: protein-coding gene | Cytoband: 8q13.1 | ||
| Entrez ID: 84675 | HGNC ID: HGNC:14215 | Ensembl Gene: ENSG00000147573 | OMIM ID: 606469 |
| Drug and gene relationship at DGIdb | |||
Expression of TRIM55:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM55 | 84675 | 232721_at | 0.1100 | 0.6429 | |
| GSE26886 | TRIM55 | 84675 | 232721_at | 0.0505 | 0.6491 | |
| GSE45670 | TRIM55 | 84675 | 232721_at | 0.1117 | 0.0739 | |
| GSE53622 | TRIM55 | 84675 | 89378 | 0.1339 | 0.3981 | |
| GSE53624 | TRIM55 | 84675 | 89378 | 0.4431 | 0.0008 | |
| GSE63941 | TRIM55 | 84675 | 236175_at | 0.0040 | 0.9885 | |
| GSE77861 | TRIM55 | 84675 | 232721_at | -0.0826 | 0.3091 | |
| GSE97050 | TRIM55 | 84675 | A_23_P123402 | -0.0118 | 0.9687 | |
| TCGA | TRIM55 | 84675 | RNAseq | 0.0513 | 0.9622 |
Upregulated datasets: 0; Downregulated datasets: 0.
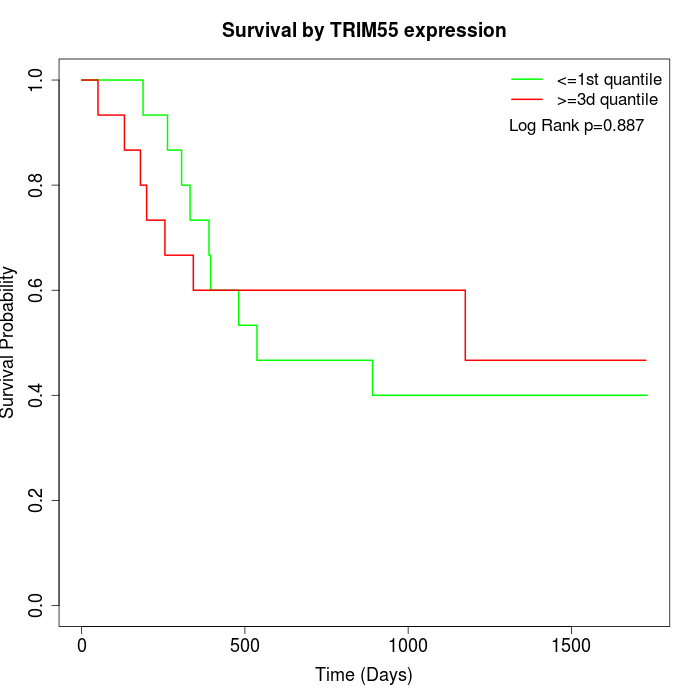
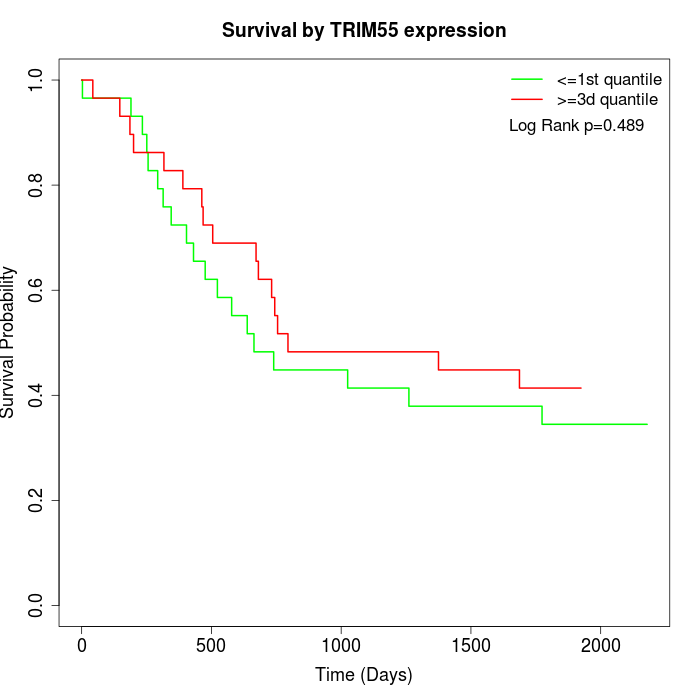
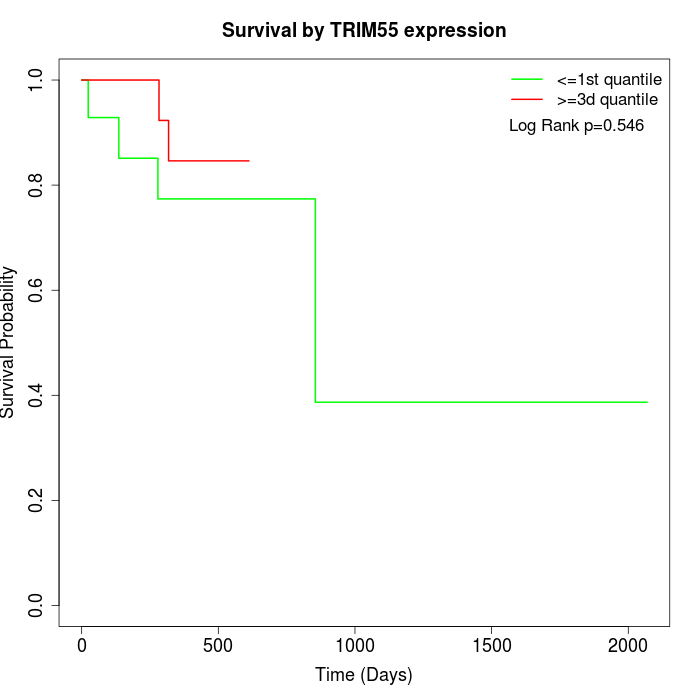
Survival by TRIM55 expression:
Note: Click image to view full size file.
Copy number change of TRIM55:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM55 | 84675 | 13 | 1 | 16 | |
| GSE20123 | TRIM55 | 84675 | 13 | 1 | 16 | |
| GSE43470 | TRIM55 | 84675 | 15 | 3 | 25 | |
| GSE46452 | TRIM55 | 84675 | 21 | 1 | 37 | |
| GSE47630 | TRIM55 | 84675 | 23 | 1 | 16 | |
| GSE54993 | TRIM55 | 84675 | 1 | 18 | 51 | |
| GSE54994 | TRIM55 | 84675 | 30 | 0 | 23 | |
| GSE60625 | TRIM55 | 84675 | 0 | 4 | 7 | |
| GSE74703 | TRIM55 | 84675 | 13 | 2 | 21 | |
| GSE74704 | TRIM55 | 84675 | 9 | 0 | 11 | |
| TCGA | TRIM55 | 84675 | 49 | 5 | 42 |
Total number of gains: 187; Total number of losses: 36; Total Number of normals: 265.
Somatic mutations of TRIM55:
Generating mutation plots.
Highly correlated genes for TRIM55:
Showing all 20 correlated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM55 | PGC | 0.743708 | 3 | 0 | 3 |
| TRIM55 | SYT3 | 0.728155 | 3 | 0 | 3 |
| TRIM55 | BTNL9 | 0.687993 | 3 | 0 | 3 |
| TRIM55 | GPR180 | 0.669528 | 3 | 0 | 3 |
| TRIM55 | OTUD4 | 0.637167 | 3 | 0 | 3 |
| TRIM55 | ZBTB17 | 0.627643 | 4 | 0 | 3 |
| TRIM55 | WFDC6 | 0.62359 | 4 | 0 | 3 |
| TRIM55 | HAPLN3 | 0.610345 | 4 | 0 | 3 |
| TRIM55 | PRR7 | 0.608394 | 3 | 0 | 3 |
| TRIM55 | LIMD1 | 0.607547 | 4 | 0 | 3 |
| TRIM55 | PLXNA3 | 0.599298 | 4 | 0 | 3 |
| TRIM55 | SH2D5 | 0.59445 | 3 | 0 | 3 |
| TRIM55 | TBX19 | 0.593592 | 4 | 0 | 3 |
| TRIM55 | UBXN11 | 0.590443 | 3 | 0 | 3 |
| TRIM55 | MED16 | 0.588328 | 3 | 0 | 3 |
| TRIM55 | TNF | 0.566383 | 4 | 0 | 3 |
| TRIM55 | CTNS | 0.562722 | 3 | 0 | 3 |
| TRIM55 | GOLGA6L6 | 0.533366 | 5 | 0 | 3 |
| TRIM55 | MEPE | 0.532949 | 6 | 0 | 3 |
| TRIM55 | GP5 | 0.521504 | 4 | 0 | 3 |
For details and further investigation, click here