| Full name: potassium two pore domain channel subfamily K member 17 | Alias Symbol: K2p17.1|TALK-2|TALK2|TASK4|TASK-4 | ||
| Type: protein-coding gene | Cytoband: 6p21.2 | ||
| Entrez ID: 89822 | HGNC ID: HGNC:14465 | Ensembl Gene: ENSG00000124780 | OMIM ID: 607370 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNK17:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNK17 | 89822 | 224049_at | -0.1551 | 0.6110 | |
| GSE26886 | KCNK17 | 89822 | 224049_at | -0.0134 | 0.9413 | |
| GSE45670 | KCNK17 | 89822 | 224049_at | -0.0555 | 0.6824 | |
| GSE53622 | KCNK17 | 89822 | 89332 | 0.0611 | 0.6494 | |
| GSE53624 | KCNK17 | 89822 | 89332 | 0.3660 | 0.0015 | |
| GSE63941 | KCNK17 | 89822 | 224049_at | 0.2325 | 0.0738 | |
| GSE77861 | KCNK17 | 89822 | 224049_at | -0.1530 | 0.1604 | |
| GSE97050 | KCNK17 | 89822 | A_23_P42128 | 0.0933 | 0.6398 | |
| SRP133303 | KCNK17 | 89822 | RNAseq | -0.1619 | 0.5583 | |
| SRP159526 | KCNK17 | 89822 | RNAseq | 0.4266 | 0.6512 | |
| TCGA | KCNK17 | 89822 | RNAseq | -0.6412 | 0.1917 |
Upregulated datasets: 0; Downregulated datasets: 0.
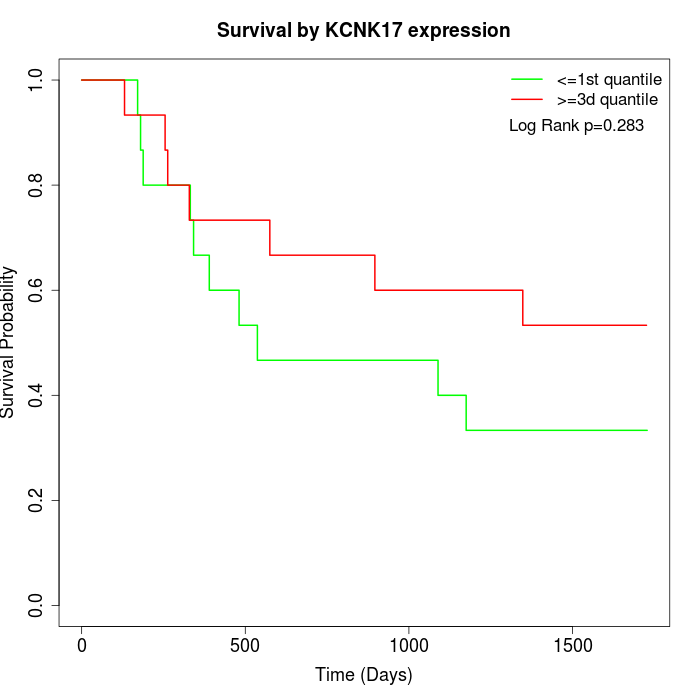
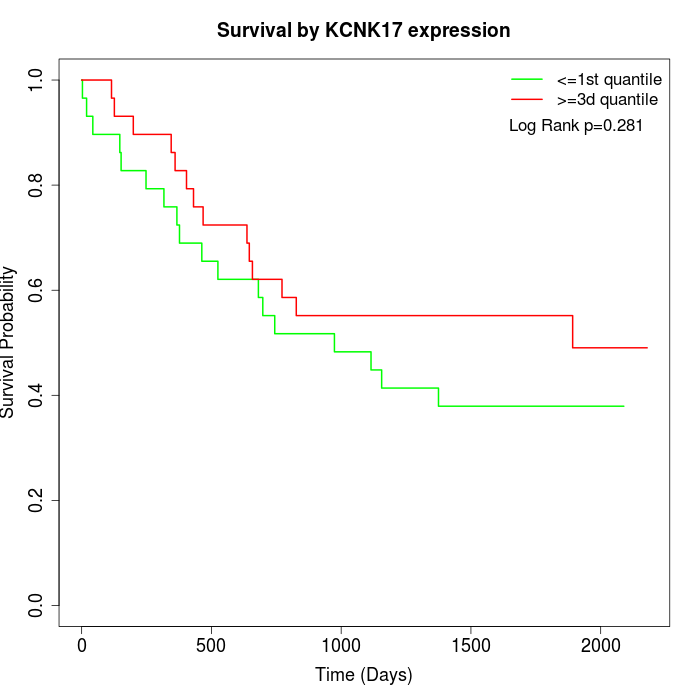
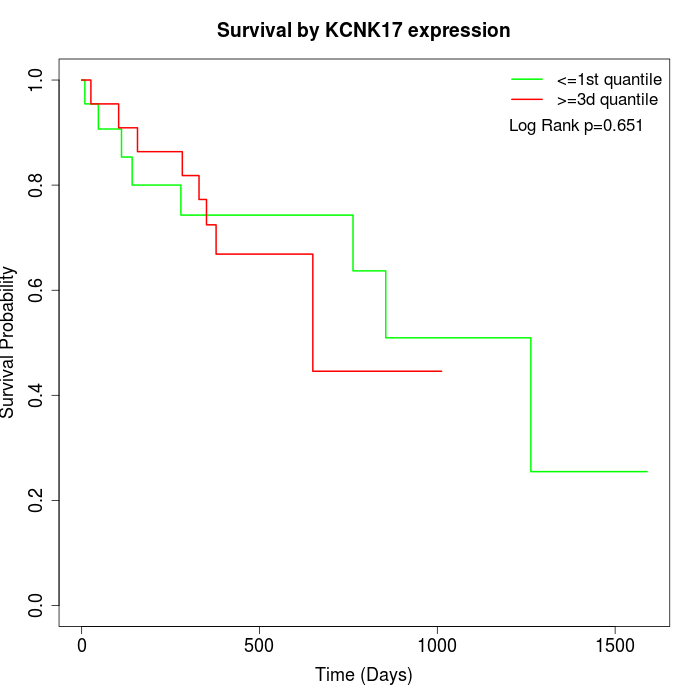
Survival by KCNK17 expression:
Note: Click image to view full size file.
Copy number change of KCNK17:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNK17 | 89822 | 4 | 1 | 25 | |
| GSE20123 | KCNK17 | 89822 | 4 | 1 | 25 | |
| GSE43470 | KCNK17 | 89822 | 6 | 0 | 37 | |
| GSE46452 | KCNK17 | 89822 | 2 | 9 | 48 | |
| GSE47630 | KCNK17 | 89822 | 8 | 4 | 28 | |
| GSE54993 | KCNK17 | 89822 | 3 | 2 | 65 | |
| GSE54994 | KCNK17 | 89822 | 11 | 4 | 38 | |
| GSE60625 | KCNK17 | 89822 | 0 | 1 | 10 | |
| GSE74703 | KCNK17 | 89822 | 6 | 0 | 30 | |
| GSE74704 | KCNK17 | 89822 | 1 | 1 | 18 | |
| TCGA | KCNK17 | 89822 | 17 | 14 | 65 |
Total number of gains: 62; Total number of losses: 37; Total Number of normals: 389.
Somatic mutations of KCNK17:
Generating mutation plots.
Highly correlated genes for KCNK17:
Showing top 20/163 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNK17 | GCNT2 | 0.771354 | 3 | 0 | 3 |
| KCNK17 | PCBP3 | 0.766247 | 3 | 0 | 3 |
| KCNK17 | ANO8 | 0.758983 | 3 | 0 | 3 |
| KCNK17 | LIPF | 0.735957 | 3 | 0 | 3 |
| KCNK17 | CCIN | 0.724921 | 3 | 0 | 3 |
| KCNK17 | B3GNT4 | 0.719412 | 3 | 0 | 3 |
| KCNK17 | ACAP2 | 0.716172 | 3 | 0 | 3 |
| KCNK17 | ATG4A | 0.712249 | 3 | 0 | 3 |
| KCNK17 | LSM12 | 0.709141 | 3 | 0 | 3 |
| KCNK17 | MCHR2 | 0.708373 | 3 | 0 | 3 |
| KCNK17 | ADAM11 | 0.707054 | 4 | 0 | 4 |
| KCNK17 | RASSF9 | 0.706716 | 3 | 0 | 3 |
| KCNK17 | FAM131B | 0.706558 | 3 | 0 | 3 |
| KCNK17 | UCP1 | 0.705871 | 4 | 0 | 4 |
| KCNK17 | ACADVL | 0.700196 | 3 | 0 | 3 |
| KCNK17 | RAB11A | 0.698701 | 3 | 0 | 3 |
| KCNK17 | BEGAIN | 0.69664 | 3 | 0 | 3 |
| KCNK17 | GIT1 | 0.68372 | 3 | 0 | 3 |
| KCNK17 | ESR2 | 0.682836 | 3 | 0 | 3 |
| KCNK17 | TSPO | 0.682399 | 3 | 0 | 3 |
For details and further investigation, click here