| Full name: protamine 2 | Alias Symbol: CT94.2 | ||
| Type: protein-coding gene | Cytoband: 16p13.13 | ||
| Entrez ID: 5620 | HGNC ID: HGNC:9448 | Ensembl Gene: ENSG00000122304 | OMIM ID: 182890 |
| Drug and gene relationship at DGIdb | |||
Expression of PRM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRM2 | 5620 | 210122_at | 0.0617 | 0.8494 | |
| GSE20347 | PRM2 | 5620 | 210122_at | -0.1778 | 0.0079 | |
| GSE23400 | PRM2 | 5620 | 210122_at | -0.2041 | 0.0000 | |
| GSE26886 | PRM2 | 5620 | 210122_at | 0.0816 | 0.4952 | |
| GSE29001 | PRM2 | 5620 | 210122_at | -0.1951 | 0.0448 | |
| GSE38129 | PRM2 | 5620 | 210122_at | -0.1692 | 0.0268 | |
| GSE45670 | PRM2 | 5620 | 210122_at | -0.0590 | 0.5999 | |
| GSE63941 | PRM2 | 5620 | 210122_at | 0.1704 | 0.1961 | |
| GSE77861 | PRM2 | 5620 | 210122_at | -0.1733 | 0.1476 |
Upregulated datasets: 0; Downregulated datasets: 0.
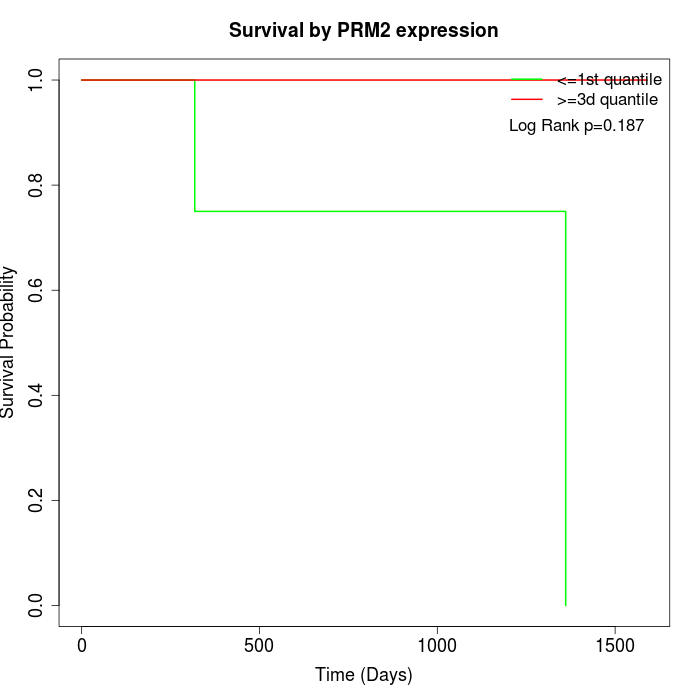
Survival by PRM2 expression:
Note: Click image to view full size file.
Copy number change of PRM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PRM2 | 5620 | 5 | 4 | 21 | |
| GSE20123 | PRM2 | 5620 | 5 | 3 | 22 | |
| GSE43470 | PRM2 | 5620 | 3 | 3 | 37 | |
| GSE46452 | PRM2 | 5620 | 38 | 1 | 20 | |
| GSE47630 | PRM2 | 5620 | 14 | 6 | 20 | |
| GSE54993 | PRM2 | 5620 | 3 | 5 | 62 | |
| GSE54994 | PRM2 | 5620 | 5 | 9 | 39 | |
| GSE60625 | PRM2 | 5620 | 4 | 0 | 7 | |
| GSE74703 | PRM2 | 5620 | 3 | 2 | 31 | |
| GSE74704 | PRM2 | 5620 | 2 | 1 | 17 | |
| TCGA | PRM2 | 5620 | 20 | 12 | 64 |
Total number of gains: 102; Total number of losses: 46; Total Number of normals: 340.
Somatic mutations of PRM2:
Generating mutation plots.
Highly correlated genes for PRM2:
Showing top 20/532 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRM2 | THRB | 0.696933 | 3 | 0 | 3 |
| PRM2 | ARSF | 0.682746 | 5 | 0 | 5 |
| PRM2 | KCNG2 | 0.669362 | 6 | 0 | 4 |
| PRM2 | OR1G1 | 0.667232 | 3 | 0 | 3 |
| PRM2 | SLC22A13 | 0.665471 | 7 | 0 | 6 |
| PRM2 | GUCY2F | 0.65182 | 3 | 0 | 3 |
| PRM2 | CT62 | 0.650212 | 3 | 0 | 3 |
| PRM2 | SLC1A7 | 0.642582 | 5 | 0 | 4 |
| PRM2 | RARG | 0.641622 | 6 | 0 | 6 |
| PRM2 | HBZ | 0.640531 | 5 | 0 | 5 |
| PRM2 | MAPK3 | 0.638971 | 6 | 0 | 5 |
| PRM2 | IL17RC | 0.636276 | 6 | 0 | 6 |
| PRM2 | CD5 | 0.631958 | 6 | 0 | 5 |
| PRM2 | SPATA25 | 0.63121 | 3 | 0 | 3 |
| PRM2 | SPTBN2 | 0.630803 | 6 | 0 | 5 |
| PRM2 | CYP3A43 | 0.629941 | 6 | 0 | 6 |
| PRM2 | DLGAP2 | 0.628923 | 4 | 0 | 4 |
| PRM2 | COX6A2 | 0.62869 | 5 | 0 | 4 |
| PRM2 | NPEPL1 | 0.627891 | 5 | 0 | 4 |
| PRM2 | TSSK2 | 0.625108 | 6 | 0 | 4 |
For details and further investigation, click here