| Full name: MIS12 kinetochore complex component | Alias Symbol: MGC2488|hMIS12|KNTC2AP|MTW1 | ||
| Type: protein-coding gene | Cytoband: 17p13.2 | ||
| Entrez ID: 79003 | HGNC ID: HGNC:24967 | Ensembl Gene: ENSG00000167842 | OMIM ID: 609178 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of MIS12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MIS12 | 79003 | 221559_s_at | 0.4394 | 0.0860 | |
| GSE20347 | MIS12 | 79003 | 221559_s_at | 0.5891 | 0.0002 | |
| GSE23400 | MIS12 | 79003 | 221559_s_at | 0.3704 | 0.0000 | |
| GSE26886 | MIS12 | 79003 | 221559_s_at | 0.2099 | 0.3902 | |
| GSE29001 | MIS12 | 79003 | 221559_s_at | 0.5804 | 0.0204 | |
| GSE38129 | MIS12 | 79003 | 221559_s_at | 0.6354 | 0.0000 | |
| GSE45670 | MIS12 | 79003 | 221559_s_at | 0.5998 | 0.0005 | |
| GSE53622 | MIS12 | 79003 | 52871 | 0.7004 | 0.0000 | |
| GSE53624 | MIS12 | 79003 | 52871 | 0.7189 | 0.0000 | |
| GSE63941 | MIS12 | 79003 | 221559_s_at | 0.5609 | 0.1330 | |
| GSE77861 | MIS12 | 79003 | 221559_s_at | 0.4340 | 0.1550 | |
| GSE97050 | MIS12 | 79003 | A_23_P78152 | 0.3991 | 0.2747 | |
| SRP007169 | MIS12 | 79003 | RNAseq | 0.5449 | 0.2358 | |
| SRP008496 | MIS12 | 79003 | RNAseq | 0.7536 | 0.0317 | |
| SRP064894 | MIS12 | 79003 | RNAseq | 0.2229 | 0.2264 | |
| SRP133303 | MIS12 | 79003 | RNAseq | 0.5455 | 0.0000 | |
| SRP159526 | MIS12 | 79003 | RNAseq | 0.4135 | 0.0281 | |
| SRP193095 | MIS12 | 79003 | RNAseq | 0.2026 | 0.0739 | |
| SRP219564 | MIS12 | 79003 | RNAseq | 0.5417 | 0.0850 | |
| TCGA | MIS12 | 79003 | RNAseq | 0.0233 | 0.6788 |
Upregulated datasets: 0; Downregulated datasets: 0.
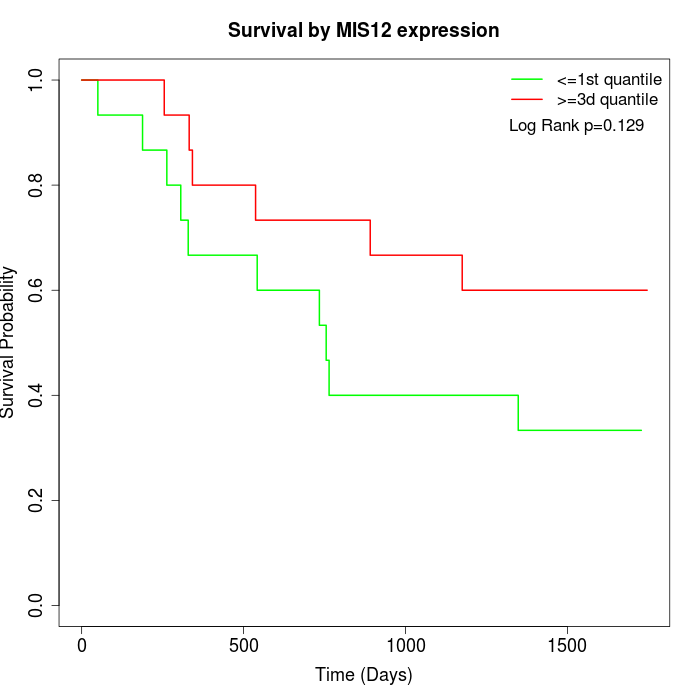
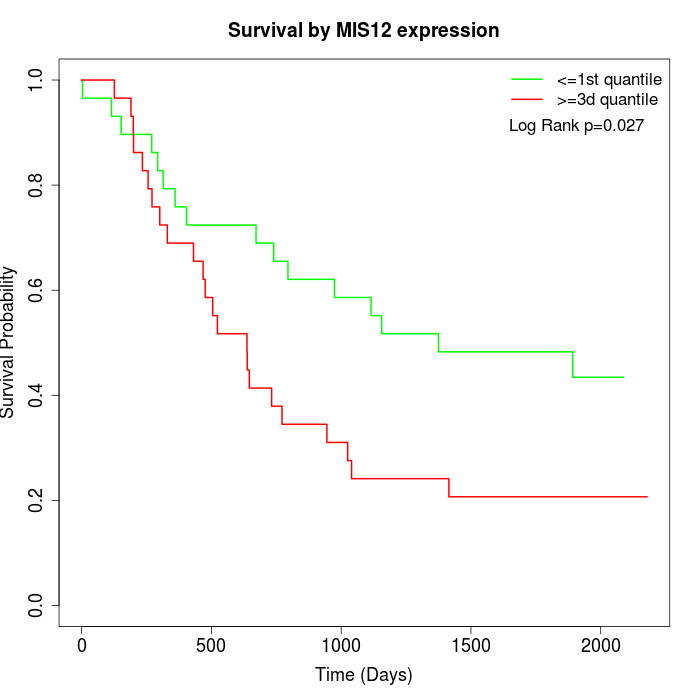
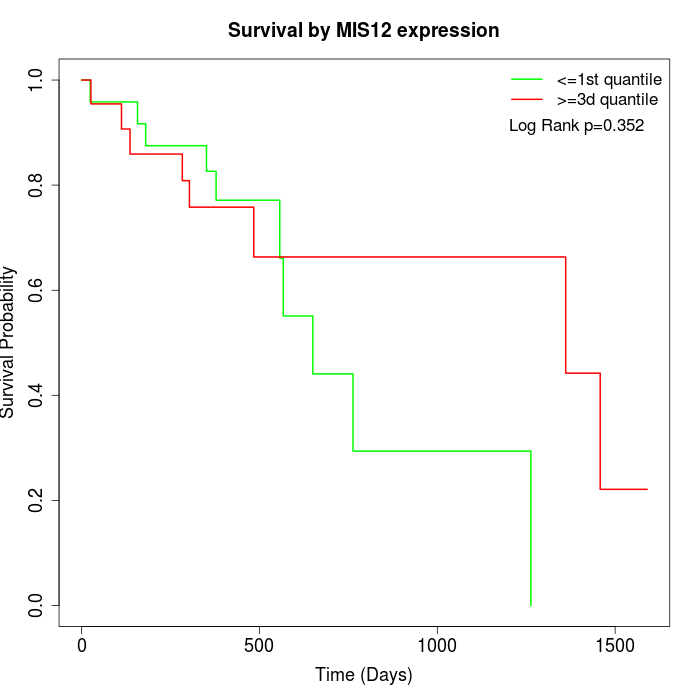
Survival by MIS12 expression:
Note: Click image to view full size file.
Copy number change of MIS12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MIS12 | 79003 | 5 | 2 | 23 | |
| GSE20123 | MIS12 | 79003 | 5 | 3 | 22 | |
| GSE43470 | MIS12 | 79003 | 1 | 6 | 36 | |
| GSE46452 | MIS12 | 79003 | 34 | 1 | 24 | |
| GSE47630 | MIS12 | 79003 | 7 | 1 | 32 | |
| GSE54993 | MIS12 | 79003 | 4 | 3 | 63 | |
| GSE54994 | MIS12 | 79003 | 5 | 8 | 40 | |
| GSE60625 | MIS12 | 79003 | 4 | 0 | 7 | |
| GSE74703 | MIS12 | 79003 | 1 | 3 | 32 | |
| GSE74704 | MIS12 | 79003 | 3 | 1 | 16 | |
| TCGA | MIS12 | 79003 | 18 | 20 | 58 |
Total number of gains: 87; Total number of losses: 48; Total Number of normals: 353.
Somatic mutations of MIS12:
Generating mutation plots.
Highly correlated genes for MIS12:
Showing top 20/1549 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MIS12 | RNF4 | 0.762887 | 3 | 0 | 3 |
| MIS12 | USP32 | 0.756998 | 3 | 0 | 3 |
| MIS12 | YWHAG | 0.753768 | 4 | 0 | 4 |
| MIS12 | ETV6 | 0.75271 | 3 | 0 | 3 |
| MIS12 | ALG1 | 0.739941 | 3 | 0 | 3 |
| MIS12 | MMS22L | 0.73864 | 3 | 0 | 3 |
| MIS12 | ATM | 0.738114 | 3 | 0 | 3 |
| MIS12 | ERP27 | 0.737825 | 3 | 0 | 3 |
| MIS12 | CWF19L1 | 0.732632 | 4 | 0 | 4 |
| MIS12 | C1orf226 | 0.730062 | 4 | 0 | 4 |
| MIS12 | FCGR3A | 0.727038 | 3 | 0 | 3 |
| MIS12 | RBM41 | 0.722673 | 3 | 0 | 3 |
| MIS12 | CENPL | 0.722031 | 6 | 0 | 6 |
| MIS12 | MAGOH | 0.717619 | 3 | 0 | 3 |
| MIS12 | C1orf131 | 0.713935 | 7 | 0 | 6 |
| MIS12 | WDR89 | 0.713196 | 4 | 0 | 4 |
| MIS12 | DHX33 | 0.712944 | 8 | 0 | 8 |
| MIS12 | TMEM138 | 0.709527 | 6 | 0 | 6 |
| MIS12 | TOP1MT | 0.704023 | 4 | 0 | 4 |
| MIS12 | UCK2 | 0.69986 | 3 | 0 | 3 |
For details and further investigation, click here