| Full name: ALG1 chitobiosyldiphosphodolichol beta-mannosyltransferase | Alias Symbol: HMT-1|HMAT1|CDG1K|Mat-1 | ||
| Type: protein-coding gene | Cytoband: 16p13.3 | ||
| Entrez ID: 56052 | HGNC ID: HGNC:18294 | Ensembl Gene: ENSG00000033011 | OMIM ID: 605907 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of ALG1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ALG1 | 56052 | 223355_at | 1.0168 | 0.0900 | |
| GSE26886 | ALG1 | 56052 | 223355_at | 0.5397 | 0.0004 | |
| GSE45670 | ALG1 | 56052 | 223355_at | 0.7070 | 0.0000 | |
| GSE63941 | ALG1 | 56052 | 223355_at | 0.0353 | 0.9437 | |
| GSE77861 | ALG1 | 56052 | 223355_at | 0.2449 | 0.0943 | |
| GSE97050 | ALG1 | 56052 | A_32_P169131 | 0.3256 | 0.2196 | |
| SRP007169 | ALG1 | 56052 | RNAseq | 0.3761 | 0.3343 | |
| SRP008496 | ALG1 | 56052 | RNAseq | 0.3318 | 0.1481 | |
| SRP064894 | ALG1 | 56052 | RNAseq | 0.7038 | 0.0015 | |
| SRP133303 | ALG1 | 56052 | RNAseq | 0.5665 | 0.0001 | |
| SRP159526 | ALG1 | 56052 | RNAseq | 0.3787 | 0.1399 | |
| SRP193095 | ALG1 | 56052 | RNAseq | 0.3253 | 0.0057 | |
| SRP219564 | ALG1 | 56052 | RNAseq | 0.7326 | 0.0432 | |
| TCGA | ALG1 | 56052 | RNAseq | -0.0973 | 0.1033 |
Upregulated datasets: 0; Downregulated datasets: 0.
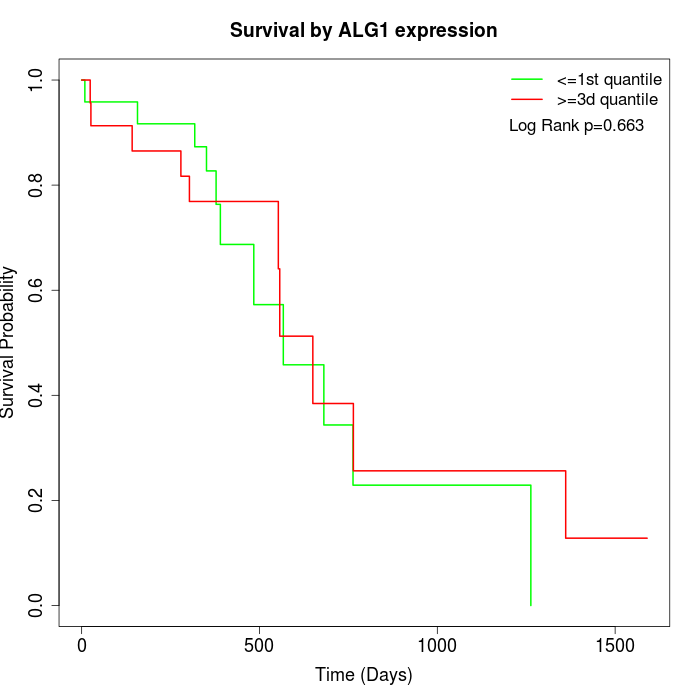
Survival by ALG1 expression:
Note: Click image to view full size file.
Copy number change of ALG1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ALG1 | 56052 | 5 | 4 | 21 | |
| GSE20123 | ALG1 | 56052 | 6 | 3 | 21 | |
| GSE43470 | ALG1 | 56052 | 4 | 4 | 35 | |
| GSE46452 | ALG1 | 56052 | 38 | 1 | 20 | |
| GSE47630 | ALG1 | 56052 | 13 | 6 | 21 | |
| GSE54993 | ALG1 | 56052 | 3 | 5 | 62 | |
| GSE54994 | ALG1 | 56052 | 5 | 9 | 39 | |
| GSE60625 | ALG1 | 56052 | 4 | 0 | 7 | |
| GSE74703 | ALG1 | 56052 | 4 | 4 | 28 | |
| GSE74704 | ALG1 | 56052 | 3 | 2 | 15 | |
| TCGA | ALG1 | 56052 | 19 | 13 | 64 |
Total number of gains: 104; Total number of losses: 51; Total Number of normals: 333.
Somatic mutations of ALG1:
Generating mutation plots.
Highly correlated genes for ALG1:
Showing top 20/1160 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ALG1 | EARS2 | 0.833581 | 3 | 0 | 3 |
| ALG1 | NPRL3 | 0.822453 | 3 | 0 | 3 |
| ALG1 | UMPS | 0.815732 | 4 | 0 | 4 |
| ALG1 | NOLC1 | 0.814486 | 3 | 0 | 3 |
| ALG1 | AGMAT | 0.809339 | 3 | 0 | 3 |
| ALG1 | INTS9 | 0.807221 | 3 | 0 | 3 |
| ALG1 | BAG4 | 0.804651 | 3 | 0 | 3 |
| ALG1 | FAM86C1 | 0.799094 | 3 | 0 | 3 |
| ALG1 | CCT6A | 0.797584 | 3 | 0 | 3 |
| ALG1 | TRAF3 | 0.792494 | 3 | 0 | 3 |
| ALG1 | AURKAIP1 | 0.792201 | 3 | 0 | 3 |
| ALG1 | KDM1A | 0.791883 | 3 | 0 | 3 |
| ALG1 | MRPL47 | 0.787368 | 5 | 0 | 5 |
| ALG1 | ZBTB9 | 0.787185 | 3 | 0 | 3 |
| ALG1 | DARS2 | 0.784445 | 3 | 0 | 3 |
| ALG1 | PRPS2 | 0.780377 | 3 | 0 | 3 |
| ALG1 | CDK6 | 0.780289 | 3 | 0 | 3 |
| ALG1 | NEK2 | 0.778605 | 3 | 0 | 3 |
| ALG1 | CSE1L | 0.778528 | 4 | 0 | 4 |
| ALG1 | MFHAS1 | 0.778294 | 5 | 0 | 5 |
For details and further investigation, click here